 |
PDBsum entry 2g1h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.39
- [acyl-carrier-protein] S-malonyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[ACP] + malonyl-CoA = malonyl-[ACP] + CoA
|
 |
 |
 |
 |
 |
holo-[ACP]
|
+
|
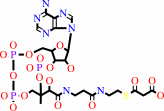 malonyl-CoA
malonyl-CoA
|
=
|
malonyl-[ACP]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:613-618
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography.
|
|
C.Oefner,
H.Schulz,
A.D'Arcy,
G.E.Dale.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Malonyl-CoA-acyl carrier protein transacylase (FabD; EC 2.3.1.39) is a key
enzyme in the fatty-acid biosynthesis pathway of bacteria, catalyzing the
transfer of a malonyl moiety from malonyl-CoA to holo acyl carrier protein
(ACP), generating malonyl-ACP and free CoASH. Malonyl-ACP, which is the product
of this reaction, is the key building block for de novo fatty-acid biosynthesis.
Various binary complex structures of the Escherichia coli enzyme are presented,
including that of the natural substrate malonyl-CoA, indicating the functional
role of the highly conserved amino acids Gln11, Ser92, Arg117 and His201 and the
stabilizing function of the preformed oxyanion hole during the enzymatic
reaction. Based on the presented structural data, a possible new catalytic
enzyme mechanism is discussed. The data obtained could be used in aiding the
process of rational inhibitor design.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Malonyl-CoA binding to FabdD. The malonyl moiety of the
processed substrate is covalently bound to the catalytic serine
residue 92 via an oxo-ester bond. Hydrogen bonding is indicated
by dotted lines. The figure was generated with the programs
MOLSCRIPT (Kraulis, 1991[Kraulis, P. J. (1991). J. Appl. Cryst.
24, 946-950.]) and RASTER3D (Merrit & Bacon, 1997[Merritt, E. A.
& Bacon, D. J. (1997). Methods Enzymol. 277, 505-524.]).
|
 |
Figure 4.
Figure 4 Reaction mechanism.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
613-618)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Kenanov,
C.Kaleta,
A.Petzold,
C.Hoischen,
S.Diekmann,
R.A.Siddiqui,
and
S.Schuster
(2010).
Theoretical study of lipid biosynthesis in wild-type Escherichia coli and in a protoplast-type L-form using elementary flux mode analysis.
|
| |
FEBS J,
277,
1023-1034.
|
 |
|
|
|
|
 |
T.Maier,
M.Leibundgut,
D.Boehringer,
and
N.Ban
(2010).
Structure and function of eukaryotic fatty acid synthases.
|
| |
Q Rev Biophys,
43,
373-422.
|
 |
|
|
|
|
 |
Y.Shen,
J.Liu,
G.Estiu,
B.Isin,
Y.Y.Ahn,
D.S.Lee,
A.L.Barabási,
V.Kapatral,
O.Wiest,
and
Z.N.Oltvai
(2010).
Blueprint for antimicrobial hit discovery targeting metabolic networks.
|
| |
Proc Natl Acad Sci U S A,
107,
1082-1087.
|
 |
|
|
|
|
 |
A.Misra,
N.Surolia,
and
A.Surolia
(2009).
Catalysis and mechanism of malonyl transferase activity in type II fatty acid biosynthesis acyl carrier proteins.
|
| |
Mol Biosyst,
5,
651-659.
|
 |
|
|
|
|
 |
E.J.Brignole,
S.Smith,
and
F.J.Asturias
(2009).
Conformational flexibility of metazoan fatty acid synthase enables catalysis.
|
| |
Nat Struct Mol Biol,
16,
190-197.
|
 |
|
|
|
|
 |
L.Kizer,
D.J.Pitera,
B.F.Pfleger,
and
J.D.Keasling
(2008).
Application of functional genomics to pathway optimization for increased isoprenoid production.
|
| |
Appl Environ Microbiol,
74,
3229-3241.
|
 |
|
|
|
|
 |
M.Leibundgut,
T.Maier,
S.Jenni,
and
N.Ban
(2008).
The multienzyme architecture of eukaryotic fatty acid synthases.
|
| |
Curr Opin Struct Biol,
18,
714-725.
|
 |
|
|
|
|
 |
T.Maier,
M.Leibundgut,
and
N.Ban
(2008).
The crystal structure of a mammalian fatty acid synthase.
|
| |
Science,
321,
1315-1322.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Ghadbane,
A.K.Brown,
L.Kremer,
G.S.Besra,
and
K.Fütterer
(2007).
Structure of Mycobacterium tuberculosis mtFabD, a malonyl-CoA:acyl carrier protein transacylase (MCAT).
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
831-835.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.B.Lomakin,
Y.Xiong,
and
T.A.Steitz
(2007).
The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together.
|
| |
Cell,
129,
319-332.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Zhang,
W.Liu,
J.Xiao,
T.Hu,
J.Chen,
K.Chen,
H.Jiang,
and
X.Shen
(2007).
Malonyl-CoA: acyl carrier protein transacylase from Helicobacter pylori: Crystal structure and its interaction with acyl carrier protein.
|
| |
Protein Sci,
16,
1184-1192.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Jenni,
M.Leibundgut,
D.Boehringer,
C.Frick,
B.Mikolásek,
and
N.Ban
(2007).
Structure of fungal fatty acid synthase and implications for iterative substrate shuttling.
|
| |
Science,
316,
254-261.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

