
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of the catalytic domain of bovine beta1,4- galactosyltransferase-i in complex with alpha-lactalbumin, ca and udp-galactose
|
|
Structure:
|
 |
Alpha-lactalbumin. Chain: a, c. Synonym: lactose synthase b protein. Engineered: yes. Mutation: yes. Beta-1,4-galactosyltransferase. Chain: b, d. Fragment: residues 57-329. Engineered: yes
|
|
Source:
|
 |
Mus musculus. House mouse. Organism_taxid: 10090. Gene: lalba. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693. Bos taurus. Cattle. Organism_taxid: 9913.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.200
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
B.Ramakrishnan,V.Ramasamy,P.K.Qasba
|
Key ref:

|
 |
B.Ramakrishnan
et al.
(2006).
Structural snapshots of beta-1,4-galactosyltransferase-I along the kinetic pathway.
J Mol Biol,
357,
1619-1633.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Feb-06
|
Release date:
|
14-Mar-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains B, D:
E.C.2.4.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains B, D:
E.C.2.4.1.22
- lactose synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + UDP-alpha-D-galactose = lactose + UDP + H+
|
 |
 |
 |
 |
 |
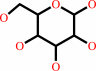 D-glucose
D-glucose
|
+
|
UDP-alpha-D-galactose
|
=
|
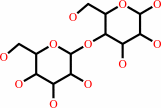 lactose
lactose
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 69.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, D:
E.C.2.4.1.275
- neolactotriaosylceramide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E)) + UDP-alpha-D-galactose = a neolactoside nLc4Cer(d18:1(4E)) + UDP + H+
|
 |
 |
 |
 |
 |
beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E))
|
+
|
UDP-alpha-D-galactose
|
=
|
neolactoside nLc4Cer(d18:1(4E))
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 69.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains B, D:
E.C.2.4.1.38
- beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acetyl-beta-D-glucosaminyl derivative + UDP-alpha-D-galactose = a beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative + UDP + H+
|
 |
 |
 |
 |
 |
N-acetyl-beta-D-glucosaminyl derivative
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 69.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains B, D:
E.C.2.4.1.90
- N-acetyllactosamine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetyl-D-glucosamine + UDP-alpha-D-galactose = beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine + UDP + H+
|
 |
 |
 |
 |
 |
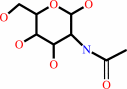 N-acetyl-D-glucosamine
N-acetyl-D-glucosamine
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
357:1619-1633
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural snapshots of beta-1,4-galactosyltransferase-I along the kinetic pathway.
|
|
B.Ramakrishnan,
V.Ramasamy,
P.K.Qasba.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
During the catalytic cycle of beta1,4-galactosyltransferase-1 (Gal-T1), upon the
binding of Mn(2+) followed by UDP-Gal, two flexible loops, a long and a short
loop, change their conformation from open to closed. We have determined the
crystal structures of a human M340H-Gal-T1 mutant in the open conformation
(apo-enzyme), its Mn(2+) and Mn(2+)-UDP-Gal-bound complexes, and of a pentenary
complex of bovine Gal-T1-Mn(2+)-UDP-GalNAc-Glc-alpha-lactalbumin. These studies
show that during the conformational changes in Gal-T1, the coordination of
Mn(2+) undergoes significant changes. It loses a coordination bond with a water
molecule bound in the open conformation of Gal-T1 while forming a new
coordination bond with another water molecule in the closed conformation,
creating an active ground-state structure that facilitates enzyme catalysis. In
the crystal structure of the pentenary complex, the N-acetylglucosamine (GlcNAc)
moiety is found cleaved from UDP-GalNAc and is placed 2.7A away from the O4
oxygen atom of the acceptor Glc molecule, yet to form the product. The anomeric
C1 atom of the cleaved GalNAc moiety has only two covalent bonds with its
non-hydrogen atoms (O5 and C2 atoms), similar to either an oxocarbenium ion or
N-acetylgalactal form, which are crystallographically indistinguishable at the
present resolution. The structure also shows that the newly formed,
metal-coordinating water molecule forms a hydrogen bond with the beta-phosphate
group of the cleaved UDP moiety. This hydrogen bond formation results in the
rotation of the beta-phosphate group of UDP away from the cleaved GalNAc moiety,
thereby preventing the re-formation of the UDP-sugar during catalysis.
Therefore, this water molecule plays an important role during catalysis in
ensuring that the catalytic reaction proceeds in a forward direction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The schematic diagram showing the kinetic pathway
of the Gal-T1 (GT) enzyme and of lactose synthase reaction
where, in the presence of aLA, glucose (Glc) is the acceptor
substrate. The crystal structures of the representative
intermediates determined here along the reaction pathway,
together with a previously determined structure, are indicated
underneath the reaction scheme with the corresponding Figures
here, in blue and red, respectively. First the apo-enzyme exists
in an open conformation (Figure 2(a)), to which the manganese
ion (Mn2+) binds (Figure 3(a)), followed by the donor substrate,
UDP-Gal (Figure 3(b)). Upon UDP-Gal or UDP-sugar binding the
enzyme undergoes conformational changes from open to closed
(Figure 3(c)), creating the acceptor and aLA binding sites. aLA
and Glc bind together synergistically to GT-Mn2+-UDP-sugar
complex in the closed conformation, forming a ground state
pentenary complex (Figure 4). During the transition state the
sugar moiety is cleaved from UDP-sugar and exists as an
oxocarbenium ion, shown as Gal* (or GalNAc* in Figure 4), which
forms a disaccharide linkage with the acceptor sugar, Glc, and
is then released from the enzyme (GT) along with the aLA
molecule from the pentenary complex. Here, we have used
UDP-GalNAc as the donor substrate to crystallize the pentenary
complex (Figure 4), since due to the steric hindrance caused by
the side-chain of Tyr286 residue with the N-acetyl moiety of
UDP-GalNAc, the transfer of GalNAc from UDP-GalNAc to Glc is
very poor, thus enabling us to crystallize the pentenary complex.
|
 |
Figure 6.
Figure 6. Metal ion-bound water molecule observed in the
crystal structures of nucleotide or sugar nucleotide-bound
complexes of other glycosyltransferases. It seems that in
Gal-T1, the presence of the metal ion-bound water molecule, W5,
is important for the rotation of the b-phosphate oxygen atoms to
form a hydrogen bond with the O1 oxygen atom, which ensures that
the catalytic reaction proceeds. Since cleavage of the sugar
moiety from the nucleotide sugar is a common step in all the
glycosyltransferases, irrespective of their catalytic mechanism,
the presence of a metal-bound water molecule in the vicinity of
the glycosidic bond of the bound nucleotide-sugar may be a
common structural feature. We have examined the (a)
b1,2-N-acetylglucosaminyltransferase (1FOA.PDB), (b)
b1,3-glucuronyltransferase I (1KWS.PDB), and (c)
a1,4-N-acetylhexosaminyltransferase (1ON6.PDB), with their
nucleotide-sugar complexes. In all these structures, a
metal-bound water molecule is found in the vicinity of the
glycosidic bond. Thus, this water molecule seems to play an
important role in the catalytic mechanism, similar to the one
(W5) found in the present crystal structures.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
357,
1619-1633)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Audry,
C.Jeanneau,
A.Imberty,
A.Harduin-Lepers,
P.Delannoy,
and
C.Breton
(2011).
Current trends in the structure-activity relationships of sialyltransferases.
|
| |
Glycobiology,
21,
716-726.
|
 |
|
|
|
|
 |
J.R.Brown,
F.Yang,
A.Sinha,
B.Ramakrishnan,
Y.Tor,
P.K.Qasba,
and
J.D.Esko
(2009).
Deoxygenated Disaccharide Analogs as Specific Inhibitors of {beta}1-4-Galactosyltransferase 1 and Selectin-mediated Tumor Metastasis.
|
| |
J Biol Chem,
284,
4952-4959.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.T.Forsee,
R.T.Cartee,
and
J.Yother
(2009).
A Kinetic Model for Chain Length Modulation of Streptococcus pneumoniae Cellubiuronan Capsular Polysaccharide by Nucleotide Sugar Donor Concentrations.
|
| |
J Biol Chem,
284,
11836-11844.
|
 |
|
|
|
|
 |
L.L.Lairson,
B.Henrissat,
G.J.Davies,
and
S.G.Withers
(2008).
Glycosyltransferases: structures, functions, and mechanisms.
|
| |
Annu Rev Biochem,
77,
521-555.
|
 |
|
|
|
|
 |
P.K.Qasba,
B.Ramakrishnan,
and
E.Boeggeman
(2008).
Structure and function of beta -1,4-galactosyltransferase.
|
| |
Curr Drug Targets,
9,
292-309.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

