 |
PDBsum entry 2fwm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2fwm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of e. Coli enta, a 2,3-dihydrodihydroxy benzoate dehydrogenase
|
|
Structure:
|
 |
2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. Chain: x. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Strain: jm109. Gene: enta. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.186
|
R-free:
|
0.214
|
|
|
Authors:
|
 |
A.M.Gulick,W.L.Duax
|
Key ref:

|
 |
J.A.Sundlov
et al.
(2006).
Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli.
Acta Crystallogr D Biol Crystallogr,
62,
734-740.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Feb-06
|
Release date:
|
27-Jun-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15047
(ENTA_ECOLI) -
2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
248 a.a.
212 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.1.28
- 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2S,3S)-2,3-dihydroxy-2,3-dihydrobenzoate + NAD+ = 2,3- dihydroxybenzoate + NADH + H+
|
 |
 |
 |
 |
 |
(2S,3S)-2,3-dihydroxy-2,3-dihydrobenzoate
|
+
|
 NAD(+)
NAD(+)
|
=
|
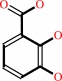 2,3- dihydroxybenzoate
2,3- dihydroxybenzoate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:734-740
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli.
|
|
J.A.Sundlov,
J.A.Garringer,
J.M.Carney,
A.S.Reger,
E.J.Drake,
W.L.Duax,
A.M.Gulick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Escherichia coli enterobactin synthetic cluster is composed of six proteins,
EntA-EntF, that form the enterobactin molecule from three serine molecules and
three molecules of 2,3-dihydroxybenzoic acid (DHB). EntC, EntB and EntA catalyze
the three-step synthesis of DHB from chorismate. EntA is a member of the
short-chain oxidoreductase (SCOR) family of proteins and catalyzes the final
step in DHB synthesis, the NAD+-dependent oxidation of
2,3-dihydro-2,3-dihydroxybenzoic acid to DHB. The structure of EntA has been
determined by multi-wavelength anomalous dispersion methods. Here, the 2.0 A
crystal structure of EntA in the unliganded form is presented. Analysis of the
structure in light of recent structural and bioinformatic analysis of other
members of the SCOR family provides insight into the residues involved in
cofactor and substrate binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Structure of the EntA monomer and tetramer. (a) Ribbon
diagram of the EntA monomer. The central  -sheet
is shown in red, while the surrounding helices are shown in
blue. The secondary-structural elements are labeled. Note that
helices -sheet
is shown in red, while the surrounding helices are shown in
blue. The secondary-structural elements are labeled. Note that
helices  1
and 1
and  3
are not present in the EntA structure; in accordance with
secondary-structure nomenclature for prior SCOR proteins (Duax
et al., 2000[Duax, W. L., Ghosh, D. & Pletnev, V. (2000). Vitam.
Horm. 58, 121-148.]), the helices are labeled 3
are not present in the EntA structure; in accordance with
secondary-structure nomenclature for prior SCOR proteins (Duax
et al., 2000[Duax, W. L., Ghosh, D. & Pletnev, V. (2000). Vitam.
Horm. 58, 121-148.]), the helices are labeled  2, 2,
 4, 4,
 5, 5,
 6,
and 6,
and  7.
(b) The EntA tetramer is shown viewed down one twofold axis of
222 crystallographic symmetry. The two top and two bottom
subunits interact around the four-helix bundle formed by helices 7.
(b) The EntA tetramer is shown viewed down one twofold axis of
222 crystallographic symmetry. The two top and two bottom
subunits interact around the four-helix bundle formed by helices
 5
and 5
and  6.
The C-terminal loops and 6.
The C-terminal loops and  7
form interactions between dimers. 7
form interactions between dimers.
|
 |
Figure 4.
Figure 4 Proposed substrate-binding pocket of EntA. The
residues of the EntA protein predicted by covariance analysis to
contribute to 2,3-DHDHB-specific binding are labeled. The
cofactor molecule (shown in orange) was modeled into the
unliganded EntA structure by superimposing the 7  -hydroxysteroid
dehydrogenase (PDB code 1fmc ) structure onto the EntA molecule. -hydroxysteroid
dehydrogenase (PDB code 1fmc ) structure onto the EntA molecule.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
734-740)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.R.Devireddy,
D.O.Hart,
D.H.Goetz,
and
M.R.Green
(2010).
A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production.
|
| |
Cell,
141,
1006-1017.
|
 |
|
|
|
|
 |
A.Koglin,
and
C.T.Walsh
(2009).
Structural insights into nonribosomal peptide enzymatic assembly lines.
|
| |
Nat Prod Rep,
26,
987.
|
 |
|
|
|
|
 |
S.A.Samel,
M.A.Marahiel,
and
L.O.Essen
(2008).
How to tailor non-ribosomal peptide products--new clues about the structures and mechanisms of modifying enzymes.
|
| |
Mol Biosyst,
4,
387-393.
|
 |
|
|
|
|
 |
E.J.Drake,
J.Cao,
J.Qu,
M.B.Shah,
R.M.Straubinger,
and
A.M.Gulick
(2007).
The 1.8 A crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa.
|
| |
J Biol Chem,
282,
20425-20434.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Miethke,
and
M.A.Marahiel
(2007).
Siderophore-based iron acquisition and pathogen control.
|
| |
Microbiol Mol Biol Rev,
71,
413-451.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

