 |
PDBsum entry 2fp0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.143
- poly(ADP-ribose) glycohydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Poly(ADP-ribose) Glycohydrolase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[(1''->2')-ADP-alpha-D-ribose](n) + H2O = [(1''->2')-ADP-alpha-D- ribose](n-1) + ADP-D-ribose
|
 |
 |
 |
 |
 |
[(1''->2')-ADP-alpha-D-ribose](n)
|
+
|
H2O
|
=
|
[(1''->2')-ADP-alpha-D- ribose](n-1)
|
+
|
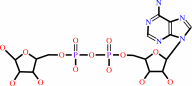 ADP-D-ribose
ADP-D-ribose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:15026-15031
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation.
|
|
C.Mueller-Dieckmann,
S.Kernstock,
M.Lisurek,
J.P.von Kries,
F.Haag,
M.S.Weiss,
F.Koch-Nolte.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Posttranslational modifications are used by cells from all kingdoms of life to
control enzymatic activity and to regulate protein function. For many cellular
processes, including DNA repair, spindle function, and apoptosis, reversible
mono- and polyADP-ribosylation constitutes a very important regulatory
mechanism. Moreover, many pathogenic bacteria secrete toxins which
ADP-ribosylate human proteins, causing diseases such as whooping cough, cholera,
and diphtheria. Whereas the 3D structures of numerous ADP-ribosylating toxins
and related mammalian enzymes have been elucidated, virtually nothing is known
about the structure of protein de-ADP-ribosylating enzymes. Here, we report the
3Dstructure of human ADP-ribosylhydrolase 3 (hARH3). The molecular architecture
of hARH3 constitutes the archetype of an all-alpha-helical protein fold and
provides insights into the reversibility of protein ADP-ribosylation. Two
magnesium ions flanked by highly conserved amino acids pinpoint the active-site
crevice. Recombinant hARH3 binds free ADP-ribose with micromolar affinity and
efficiently de-ADP-ribosylates poly- but not monoADP-ribosylated proteins.
Docking experiments indicate a possible binding mode for ADP-ribose polymers and
suggest a reaction mechanism. Our results underscore the importance of
endogenous ADP-ribosylation cycles and provide a basis for structure-based
design of ADP-ribosylhydrolase inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Posttranslational modification of proteins by
reversible ADP-ribosylation. ARTs and PARPs transfer the
ADP-ribose (ADPR) moiety from  -NAD onto specific amino
acid side chains or onto ADPR moieties (X) of target proteins
under the release of nicotinamide. This modification may lead to
either activation or inactivation of the target protein.
Protein-ADP-ribosylhydrolases (ARHs and PARGs) hydrolyze the -NAD onto specific amino
acid side chains or onto ADPR moieties (X) of target proteins
under the release of nicotinamide. This modification may lead to
either activation or inactivation of the target protein.
Protein-ADP-ribosylhydrolases (ARHs and PARGs) hydrolyze the
 -glycosidic bond
between ADPR and the side chain, thereby restoring normal
protein function. X can be Arg, Asp, Cys, diphthamide, Glu, or
ADPR. In the case of mono-ADP-ribosylation, R and R' are OH
groups. In the case of polyADP-ribosylation, attachment of ADPR
can take place at the R site (elongation) or at the R' site
(branching). In mammals, two distinct subfamilies of ARTs
(ART1–5, PARP1–17) and two distinct subfamilies of ARHs
(ARH1–3, PARG) exist. -glycosidic bond
between ADPR and the side chain, thereby restoring normal
protein function. X can be Arg, Asp, Cys, diphthamide, Glu, or
ADPR. In the case of mono-ADP-ribosylation, R and R' are OH
groups. In the case of polyADP-ribosylation, attachment of ADPR
can take place at the R site (elongation) or at the R' site
(branching). In mammals, two distinct subfamilies of ARTs
(ART1–5, PARP1–17) and two distinct subfamilies of ARHs
(ARH1–3, PARG) exist.
|
 |
Figure 3.
Fig. 3. Active site of ARH3. (A) Coordination of Mg^2+ ions
in the orthorhombic crystal form of hARH3. Hydrogen bonds are
represented as dashed lines. (B) Superposition of the
Mg^2+-coordinating residues of the orthorhombic (gray) and
monoclinic crystal forms. Residue Asp-300 is slightly shifted
but retains its bidentate binding character, whereas Glu-25 of
the monoclinic crystal form is shifted by 1.8 Å with
respect to those of the orthorhombic crystal form. The residues
from the monoclinic crystal form are shown in color.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Slade,
M.S.Dunstan,
E.Barkauskaite,
R.Weston,
P.Lafite,
N.Dixon,
M.Ahel,
D.Leys,
and
I.Ahel
(2011).
The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase.
|
| |
Nature,
477,
616-620.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.P.Strosznajder,
K.Czubowicz,
H.Jesko,
and
J.B.Strosznajder
(2010).
Poly(ADP-ribose) metabolism in brain and its role in ischemia pathology.
|
| |
Mol Neurobiol,
41,
187-196.
|
 |
|
|
|
|
 |
C.L.Berthold,
H.Wang,
S.Nordlund,
and
M.Högbom
(2009).
Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG.
|
| |
Proc Natl Acad Sci U S A,
106,
14247-14252.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Mueller-Dieckmann,
S.Kernstock,
J.Mueller-Dieckmann,
M.S.Weiss,
and
F.Koch-Nolte
(2008).
Structure of mouse ADP-ribosylhydrolase 3 (mARH3).
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
156-162.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Niere,
S.Kernstock,
F.Koch-Nolte,
and
M.Ziegler
(2008).
Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix.
|
| |
Mol Cell Biol,
28,
814-824.
|
 |
|
|
|
|
 |
O.Okhrimenko,
and
I.Jelesarov
(2008).
A survey of the year 2006 literature on applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
21,
1.
|
 |
|
|
|
|
 |
C.Mueller-Dieckmann,
S.Panjikar,
A.Schmidt,
S.Mueller,
J.Kuper,
A.Geerlof,
M.Wilmanns,
R.K.Singh,
P.A.Tucker,
and
M.S.Weiss
(2007).
On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
366-380.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

