 |
PDBsum entry 2f3d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase
|
|
Structure:
|
 |
Fructose-1,6-bisphosphatase 1. Chain: a. Synonym: d-fructose-1,6-bisphosphate 1-phosphohydrolase 1, fbpase 1. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823. Gene: fbp1, fbp. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
1.83Å
|
R-factor:
|
0.222
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
C.V.Iancu,S.Mukund,J.-Y.Choe,H.J.Fromm,R.B.Honzatko
|
|
Key ref:
|
 |
Y.Gao
et al.
(2013).
Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry,
52,
5206-5216.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
21-Nov-05
|
Release date:
|
25-Apr-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00636
(F16P1_PIG) -
Fructose-1,6-bisphosphatase 1 from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
338 a.a.
328 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
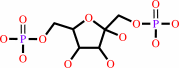 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
H2O
|
=
|
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
52:5206-5216
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
|
|
Y.Gao,
C.V.Iancu,
S.Mukind,
J.Y.Choe,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
AMP triggers a 15° subunit-pair rotation in fructose-1,6-bisphosphatase
(FBPase) from its active R state to its inactive T state. During this
transition, a catalytically essential loop (residues 50-72) leaves its active
(engaged) conformation. Here, the structures of Ile(10) → Asp FBPase and
molecular dynamic simulations reveal factors responsible for loop displacement.
The AMP/Mg(2+) and AMP/Zn(2+) complexes of Asp(10) FBPase are in intermediate
quaternary conformations (completing 12° of the subunit-pair rotation), but the
complex with Zn(2+) provides the first instance of an engaged loop in a near-T
quaternary state. The 12° subunit-pair rotation generates close contacts
involving the hinges (residues 50-57) and hairpin turns (residues 58-72) of the
engaged loops. Additional subunit-pair rotation toward the T state would make
such contacts unfavorable, presumably causing displacement of the loop. Targeted
molecular dynamics simulations reveal no steric barriers to subunit-pair
rotations of up to 14° followed by the displacement of the loop from the active
site. Principal component analysis reveals high-amplitude motions that
exacerbate steric clashes of engaged loops in the near-T state. The results of
the simulations and crystal structures are in agreement: subunit-pair rotations
just short of the canonical T state coupled with high-amplitude modes sterically
displace the dynamic loop from the active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

