 |
PDBsum entry 2exb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
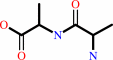
|
+
|
|
=
|
2
×
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.21.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
45:783-792
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics.
|
|
H.Kishida,
S.Unzai,
D.I.Roper,
A.Lloyd,
S.Y.Park,
J.R.Tame.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of penicillin binding protein 4 (PBP4) from Escherichia
coli, which has both DD-endopeptidase and DD-carboxypeptidase activity, is
presented. PBP4 is one of 12 penicillin binding proteins in E. coli involved in
the synthesis and maintenance of the cell wall. The model contains a penicillin
binding domain similar to known structures, but includes a large insertion which
folds into domains with unique folds. The structures of the protein covalently
attached to five different antibiotics presented here show the active site
residues are unmoved compared to the apoprotein, but nearby surface loops and
helices are displaced in some cases. The altered geometry of conserved active
site residues compared with those of other PBPs suggests a possible cause for
the slow deacylation rate of PBP4.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.I.O'Daniel,
J.Zajicek,
W.Zhang,
Q.Shi,
J.F.Fisher,
and
S.Mobashery
(2010).
Elucidation of the structure of the membrane anchor of penicillin-binding protein 5 of Escherichia coli.
|
| |
J Am Chem Soc,
132,
4110-4118.
|
 |
|
|
|
|
 |
A.J.Powell,
J.Tomberg,
A.M.Deacon,
R.A.Nicholas,
and
C.Davies
(2009).
Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance.
|
| |
J Biol Chem,
284,
1202-1212.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.C.Marciano,
N.G.Brown,
and
T.Palzkill
(2009).
Analysis of the plasticity of location of the Arg244 positive charge within the active site of the TEM-1 beta-lactamase.
|
| |
Protein Sci,
18,
2080-2089.
|
 |
|
|
|
|
 |
S.Peddi,
R.A.Nicholas,
and
W.G.Gutheil
(2009).
Neisseria gonorrhoeae penicillin-binding protein 3 demonstrates a pronounced preference for N(epsilon)-acylated substrates.
|
| |
Biochemistry,
48,
5731-5737.
|
 |
|
|
|
|
 |
E.Sauvage,
A.J.Powell,
J.Heilemann,
H.R.Josephine,
P.Charlier,
C.Davies,
and
R.F.Pratt
(2008).
Crystal structures of complexes of bacterial DD-peptidases with peptidoglycan-mimetic ligands: the substrate specificity puzzle.
|
| |
J Mol Biol,
381,
383-393.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Sauvage,
F.Kerff,
M.Terrak,
J.A.Ayala,
and
P.Charlier
(2008).
The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
234-258.
|
 |
|
|
|
|
 |
W.Vollmer,
B.Joris,
P.Charlier,
and
S.Foster
(2008).
Bacterial peptidoglycan (murein) hydrolases.
|
| |
FEMS Microbiol Rev,
32,
259-286.
|
 |
|
|
|
|
 |
I.Kumar,
H.R.Josephine,
and
R.F.Pratt
(2007).
Reactions of peptidoglycan-mimetic beta-lactams with penicillin-binding proteins in vivo and in membranes.
|
| |
ACS Chem Biol,
2,
620-624.
|
 |
|
|
|
|
 |
M.Firczuk,
and
M.Bochtler
(2007).
Folds and activities of peptidoglycan amidases.
|
| |
FEMS Microbiol Rev,
31,
676-691.
|
 |
|
|
|
|
 |
P.Macheboeuf,
C.Contreras-Martel,
V.Job,
O.Dideberg,
and
A.Dessen
(2006).
Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes.
|
| |
FEMS Microbiol Rev,
30,
673-691.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

