 |
PDBsum entry 2ev1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.6.1.1
- adenylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ATP = 3',5'-cyclic AMP + diphosphate
|
 |
 |
 |
 |
 |
 ATP
ATP
|
=
|
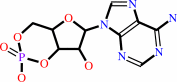 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
369:1282-1295
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid.
|
|
F.Findeisen,
J.U.Linder,
A.Schultz,
J.E.Schultz,
B.Brügger,
F.Wieland,
I.Sinning,
I.Tews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The universal secondary messenger cAMP is produced by adenylyl cyclases (ACs).
Most bacterial and all eukaryotic ACs belong to class III of six divergent
classes. A class III characteristic is formation of the catalytic pocket at a
dimer interface and the presence of additional regulatory domains. Mycobacterium
tuberculosis possesses 15 class III ACs, including Rv1264, which is activated at
acidic pH due to pH-dependent structural transitions of the Rv1264 dimer. It has
been shown by X-ray crystallography that the N-terminal regulatory and
C-terminal catalytic domains of Rv1264 interact in completely different ways in
the active and inhibited states. Here, we report an in-depth structural and
functional analysis of the regulatory domain of Rv1264. The 1.6 A resolution
crystal structure shows the protein in a tight, disk-shaped dimer, formed around
a helical bundle, and involving a protein chain crossover. To understand pH
regulation, we determined structures at acidic and basic pH values and employed
structure-based mutagenesis in the holoenzyme to elucidate regulation using an
AC activity assay. It has been shown that regulatory and catalytic domains must
be linked in a single protein chain. The new studies demonstrate that the length
of the linker segment is decisive for regulation. Several amino acids on the
surface of the regulatory domain, when exchanged, altered the pH-dependence of
AC activity. However, these residues are not conserved amongst a number of
related ACs. The closely related mycobacterial Rv2212, but not Rv1264, is
strongly activated by the addition of fatty acids. The structure resolved the
presence of a deeply embedded fatty acid, characterised as oleic acid by mass
spectrometry, which may serve as a hinge. From these data, we conclude that the
regulatory domain is a structural scaffold used for distinct regulatory purposes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Enzyme regulation requires integrity of the linker
segment that forms the αN10-switch in the Rv1264 holoenzyme.
(a) Comparison of the region forming the αN10-switch in the
active state (PDB entry 1Y11, shown in green and ochre) and the
inhibited state (PDB entry 1Y10, shown in blue and red). The
overlay is based on superposition of the regulatory domains; the
catalytic domains are offset by 55° rotation and 6 Å
movement between active and inhibited conformations.^8 The
ribbon diagrams show with helix α10 starting at residue 160,
the linker with the αN10-switch, and the first β-strand (β1)
of the catalytic domains. The position of residue 204, where
additional amino acids were inserted in the mutagenesis study,
is indicated for active and inhibited states. Positions of
residues Met193 and Met194 that have been exchanged in the
earlier study are indicated.^8 (b) pH activity profiles of
insertion of one, two, three or nine amino acids at position 204
can be seen. Standard deviations are shown as vertical bars if
they exceed the symbol size, which corresponds to SD = 10%.
While one or two inserted amino acids ((●) +S204 and (○)
+SA204) result in an overall inhibited phenotype, insertion of
three residues ((▪) +SAA204) displays a phenotype similar to
the wild-type enzyme (broken line). Figure 3. Enzyme
regulation requires integrity of the linker segment that forms
the αN10-switch in the Rv1264 holoenzyme. (a) Comparison of the
region forming the αN10-switch in the active state (PDB entry
1Y11, shown in green and ochre) and the inhibited state (PDB
entry 1Y10, shown in blue and red). The overlay is based on
superposition of the regulatory domains; the catalytic domains
are offset by 55° rotation and 6 Å movement between
active and inhibited conformations.[3]^8 The ribbon diagrams
show with helix α10 starting at residue 160, the linker with
the αN10-switch, and the first β-strand (β1) of the catalytic
domains. The position of residue 204, where additional amino
acids were inserted in the mutagenesis study, is indicated for
active and inhibited states. Positions of residues Met193 and
Met194 that have been exchanged in the earlier study are
indicated.[4]^8 (b) pH activity profiles of insertion of one,
two, three or nine amino acids at position 204 can be seen.
Standard deviations are shown as vertical bars if they exceed
the symbol size, which corresponds to SD = 10%. While one or two
inserted amino acids ((●) +S204 and (○) +SA204) result in an
overall inhibited phenotype, insertion of three residues ((▪)
+SAA204) displays a phenotype similar to the wild-type enzyme
(broken line). Introduction of the nonapeptide SAAGPSGAA ((□)
+9aa-204) leads to a decoupling of regulatory and catalytic
domains, as this enzyme is, like the M193P/M194P mutant, no
longer pH-responsive.[5]^8
|
 |
Figure 7.
Figure 7. Model for the mechanism of Rv1264 regulation,
illustrated on the active (green) and inhibited states (blue) of
the holoenzyme, shown in surface representation. The two
structural features that accompany enzyme regulation are the
extension of α10 by the αN10-switch region and the different
placement of the fatty acid molecule, accompanied by a movement
of helix α4. Figure 7. Model for the mechanism of Rv1264
regulation, illustrated on the active (green) and inhibited
states (blue) of the holoenzyme, shown in surface
representation. The two structural features that accompany
enzyme regulation are the extension of α10 by the αN10-switch
region and the different placement of the fatty acid molecule,
accompanied by a movement of helix α4.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
369,
1282-1295)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.A.Charania,
K.L.Brockman,
Y.Zhang,
A.Banerjee,
G.E.Pinchuk,
J.K.Fredrickson,
A.S.Beliaev,
and
D.A.Saffarini
(2009).
Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1.
|
| |
J Bacteriol,
191,
4298-4306.
|
 |
|
|
|
|
 |
J.U.Linder,
and
J.E.Schultz
(2008).
Versatility of signal transduction encoded in dimeric adenylyl cyclases.
|
| |
Curr Opin Struct Biol,
18,
667-672.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

