 |
PDBsum entry 2e6y

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Covalent complex of orotidine 5'-monophosphate decarboxylase (odcase) with 6-iodo-ump
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a, b. Synonym: omp decarboxylase, ompdcase, ompdecase, orotidine 5'- monophosphate decarboxylase, odcase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Methanothermobacter thermautotrophicus. Organism_taxid: 145262. Gene: pyrf. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.162
|
R-free:
|
0.187
|
|
|
Authors:
|
 |
M.Fujihashi,A.M.Bello,L.P.Kotra,E.F.Pai
|
|
Key ref:
|
 |
A.M.Bello
et al.
(2007).
A potent, covalent inhibitor of orotidine 5'-monophosphate decarboxylase with antimalarial activity.
J Med Chem,
50,
915-921.
PubMed id:
|
 |
|
Date:
|
 |
|
05-Jan-07
|
Release date:
|
27-Feb-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O26232
(PYRF_METTH) -
Orotidine 5'-phosphate decarboxylase from Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
228 a.a.
215 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
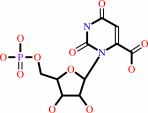 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
50:915-921
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A potent, covalent inhibitor of orotidine 5'-monophosphate decarboxylase with antimalarial activity.
|
|
A.M.Bello,
E.Poduch,
M.Fujihashi,
M.Amani,
Y.Li,
I.Crandall,
R.Hui,
P.I.Lee,
K.C.Kain,
E.F.Pai,
L.P.Kotra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Orotidine 5'-monophosphate decarboxylase (ODCase) has evolved to catalyze the
decarboxylation of orotidine 5'-monophosphate without any covalent
intermediates. Active site residues in ODCase are involved in an extensive
hydrogen-bonding network. We discovered that 6-iodouridine 5'-monophosphate
(6-iodo-UMP) irreversibly inhibits the catalytic activities of ODCases from
Methanobacterium thermoautotrophicum and Plasmodium falciparum. Mass spectral
analysis of the enzyme-inhibitor complex confirms covalent attachment of the
inhibitor to ODCase accompanied by the loss of two protons and the iodo moiety.
The X-ray crystal structure (1.6 A resolution) of the complex of the inhibitor
and ODCase clearly shows the covalent bond formation with the active site Lys-72
[corrected] residue. 6-Iodo-UMP inhibits ODCase in a time- and
concentration-dependent fashion. 6-Iodouridine, the nucleoside form of
6-iodo-UMP, exhibited potent antiplasmodial activity, with IC50s of 4.4 +/- 1.3
microM and 6.2 +/- 0.7 microM against P. falciparum ItG and 3D7 isolates,
respectively. 6-Iodouridine 5'-monophosphate is a novel covalent inhibitor of
ODCase, and its nucleoside analogue paves the way to a new class of inhibitors
against malaria.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Nencka,
D.Sinnaeve,
I.Karalic,
J.C.Martins,
and
S.Van Calenbergh
(2010).
Synthesis of C-6-substituted uridine phosphonates through aerobic ligand-free Suzuki-Miyaura cross-coupling.
|
| |
Org Biomol Chem,
8,
5234-5246.
|
 |
|
|
|
|
 |
Y.J.Wu,
C.C.Liao,
C.H.Jen,
Y.C.Shih,
and
T.C.Chien
(2010).
Chemical models and their mechanistic implications for the transformation of 6-cyanouridine 5'-monophosphate catalyzed by orotidine 5'-monophosphate decarboxylase.
|
| |
Chem Commun (Camb),
46,
4821-4823.
|
 |
|
|
|
|
 |
D.Heinrich,
U.Diederichsen,
and
M.G.Rudolph
(2009).
Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase.
|
| |
Chemistry,
15,
6619-6625.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.G.Wittmann,
D.Heinrich,
K.Gasow,
A.Frey,
U.Diederichsen,
and
M.G.Rudolph
(2008).
Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
|
| |
Structure,
16,
82-92.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Weigelt,
L.D.McBroom-Cerajewski,
M.Schapira,
Y.Zhao,
C.H.Arrowsmith,
and
C.H.Arrowmsmith
(2008).
Structural genomics and drug discovery: all in the family.
|
| |
Curr Opin Chem Biol,
12,
32-39.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

