
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.2.3.2.2
- gamma-glutamyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-terminal (5-L-glutamyl)-[peptide] + an alpha-amino acid = 5-L- glutamyl amino acid + an N-terminal L-alpha-aminoacyl-[peptide]
|
 |
 |
 |
 |
 |
N-terminal (5-L-glutamyl)-[peptide]
|
+
|
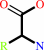 alpha-amino acid
alpha-amino acid
|
=
|
 5-L- glutamyl amino acid
5-L- glutamyl amino acid
|
+
|
N-terminal L-alpha-aminoacyl-[peptide]
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.3.4.19.13
- glutathione gamma-glutamate hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glutathione + H2O = L-cysteinylglycine + L-glutamate
|
|
2.
|
an S-substituted glutathione + H2O = an S-substituted L-cysteinylglycine + L-glutamate
|
|
 |
 |
 |
 |
 |
 glutathione
glutathione
|
+
|
H2O
|
=
|
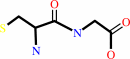 L-cysteinylglycine
L-cysteinylglycine
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
S-substituted glutathione
|
+
|
H2O
|
=
|
S-substituted L-cysteinylglycine
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:2433-2439
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the gamma-glutamyltranspeptidase precursor protein from Escherichia coli. Structural changes upon autocatalytic processing and implications for the maturation mechanism.
|
|
T.Okada,
H.Suzuki,
K.Wada,
H.Kumagai,
K.Fukuyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Gamma-glutamyltranspeptidase (GGT) is an extracellular enzyme that plays a key
role in glutathione metabolism. The mature GGT is a heterodimer consisting of L-
and S-subunits that is generated by posttranslational cleavage of the peptide
bond between Gln-390 and Thr-391 in the precursor protein. Thr-391, which
becomes the N-terminal residue of the S-subunit, acts as the active residue in
the catalytic reaction. The crystal structure of a mutant GGT, T391A, that is
unable to undergo autocatalytic processing, has been determined at 2.55-A
resolution. Structural comparison of the precursor protein and mature GGT
demonstrates that the structures of the core regions in the two proteins are
unchanged, but marked differences are found near the active site. In particular,
in the precursor, the segment corresponding to the C-terminal region of the
L-subunit occupies the site where the loop (residues 438-449) forms the lid of
the gamma-glutamyl group-binding pocket in the mature GGT. This result
demonstrates that, upon cleavage of the N-terminal peptide bond of Thr-391, the
newly produced C terminus (residues 375-390) flips out, allowing the 438-449
segment to form the gamma-glutamyl group-binding pocket. The electron density
map for the T391A protein also identified a water molecule near the carbonyl
carbon atom of Gln-390. The spatial arrangement around the water and Thr-391
relative to the scissile peptide bond appears suitable for the initiation of
autocatalytic processing, as in other members of the N-terminal nucleophile
hydrolase superfamily.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. A stereo view of the F[o] – F[c] omit map
around the processing site (A molecule). The map was generated
on the basis of F[c] calculated from the model, which was
derived from the refinement using REFMAC5 (23) omitting residues
385–392 and the water molecule (W4). The map was contoured at
the 2.5  level. A ball-and-stick
model of the T391A protein is overlaid on the map. The arrow
indicates the scissile peptide bond that is cleaved in the
wild-type precursor protein (Gln-390 to Thr-391). The figure was
prepared using PYMOL (31). level. A ball-and-stick
model of the T391A protein is overlaid on the map. The arrow
indicates the scissile peptide bond that is cleaved in the
wild-type precursor protein (Gln-390 to Thr-391). The figure was
prepared using PYMOL (31).
|
 |
Figure 2.
FIGURE 2. The tertiary structure of the T391A protein. A, a
ribbon drawing of the T391A protein (B molecule). The segments
in the T391A protein that correspond to the L- and S-subunits
are pink and green, respectively, and the P-segment (residue
375–390) is highlighted in orange. Terminal residues that
generate invisible segments are labeled. The orange arrow
indicates the site at which autocatalytic processing occurs. B,
a stereo view of the superimposition of C  traces of the T391A
protein and mature GGT. The structure of mature GGT (A molecule
of SeMet-GGT in (19)) was superimposed on that of the T391A
protein (B molecule). P-segment residues in the T391A protein
and in mature GGT are orange and blue, respectively. Residues
that had C traces of the T391A
protein and mature GGT. The structure of mature GGT (A molecule
of SeMet-GGT in (19)) was superimposed on that of the T391A
protein (B molecule). P-segment residues in the T391A protein
and in mature GGT are orange and blue, respectively. Residues
that had C  atoms displaced by >1
Å upon processing are in yellow. Residues of mature GGT
that are invisible in the T391A protein are shown in black.
Regions of invisible residues are circled in green. The distance
between Ser-387 C and Thr-391 N in mature GGT is shown. B is
rotated by 30° around the vertical axis relative to A. C, a
close-up view of the segment Glu-377 to Pro-380. A stick model
of mature GGT (blue) is superimposed on the T391A protein
(orange). The figures were prepared using PYMOL (31). atoms displaced by >1
Å upon processing are in yellow. Residues of mature GGT
that are invisible in the T391A protein are shown in black.
Regions of invisible residues are circled in green. The distance
between Ser-387 C and Thr-391 N in mature GGT is shown. B is
rotated by 30° around the vertical axis relative to A. C, a
close-up view of the segment Glu-377 to Pro-380. A stick model
of mature GGT (blue) is superimposed on the T391A protein
(orange). The figures were prepared using PYMOL (31).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
2433-2439)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Castellano,
A.Di Salle,
A.Merlino,
M.Rossi,
and
F.La Cara
(2011).
Gene cloning and protein expression of γ-glutamyltranspeptidases from Thermus thermophilus and Deinococcus radiodurans: comparison of molecular and structural properties with mesophilic counterparts.
|
| |
Extremophiles,
15,
259-270.
|
 |
|
|
|
|
 |
T.Destro,
D.Prasad,
D.Martignago,
I.L.Bernet,
A.R.Trentin,
I.K.Renu,
M.Ferretti,
and
A.Masi
(2011).
Compensatory expression and substrate inducibility of gamma-glutamyl transferase GGT2 isoform in Arabidopsis thaliana.
|
| |
J Exp Bot,
62,
805-814.
|
 |
|
|
|
|
 |
F.H.Hausheer,
D.Shanmugarajah,
B.D.Leverett,
X.Chen,
Q.Huang,
H.Kochat,
P.N.Petluru,
and
A.R.Parker
(2010).
Mechanistic study of BNP7787-mediated cisplatin nephroprotection: modulation of gamma-glutamyl transpeptidase.
|
| |
Cancer Chemother Pharmacol,
65,
941-951.
|
 |
|
|
|
|
 |
H.P.Chang,
W.C.Liang,
R.C.Lyu,
M.C.Chi,
T.F.Wang,
K.L.Su,
H.C.Hung,
and
L.L.Lin
(2010).
Effects of C-terminal truncation on autocatalytic processing of Bacillus licheniformis gamma-glutamyl transpeptidase.
|
| |
Biochemistry (Mosc),
75,
919-929.
|
 |
|
|
|
|
 |
H.Suzuki,
C.Yamada,
K.Kijima,
S.Ishihara,
K.Wada,
K.Fukuyama,
and
H.Kumagai
(2010).
Enhancement of glutaryl-7-aminocephalosporanic acid acylase activity of gamma-glutamyltranspeptidase of Bacillus subtilis.
|
| |
Biotechnol J,
5,
829-837.
|
 |
|
|
|
|
 |
K.Wada,
M.Irie,
H.Suzuki,
and
K.Fukuyama
(2010).
Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket.
|
| |
FEBS J,
277,
1000-1009.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.C.Lyu,
H.Y.Hu,
L.Y.Kuo,
H.F.Lo,
P.L.Ong,
H.P.Chang,
and
L.L.Lin
(2009).
Role of the conserved Thr399 and Thr417 residues of Bacillus licheniformis gamma-Glutamyltranspeptidase as evaluated by mutational analysis.
|
| |
Curr Microbiol,
59,
101-106.
|
 |
|
|
|
|
 |
R.Wu,
S.Richter,
R.G.Zhang,
V.J.Anderson,
D.Missiakas,
and
A.Joachimiak
(2009).
Crystal structure of Bacillus anthracis transpeptidase enzyme CapD.
|
| |
J Biol Chem,
284,
24406-24414.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Yamada,
K.Kijima,
S.Ishihara,
C.Miwa,
K.Wada,
T.Okada,
K.Fukuyama,
H.Kumagai,
and
H.Suzuki
(2008).
Improvement of the glutaryl-7-aminocephalosporanic acid acylase activity of a bacterial gamma-glutamyltranspeptidase.
|
| |
Appl Environ Microbiol,
74,
3400-3409.
|
 |
|
|
|
|
 |
O.D.Ekici,
M.Paetzel,
and
R.E.Dalbey
(2008).
Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration.
|
| |
Protein Sci,
17,
2023-2037.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

