 |
PDBsum entry 2dxl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.46
- glycerophosphodiester phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a sn-glycero-3-phosphodiester + H2O = an alcohol + sn-glycerol 3-phosphate + H+
|
 |
 |
 |
 |
 |
sn-glycero-3-phosphodiester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
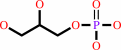 sn-glycerol 3-phosphate
sn-glycerol 3-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
367:1047-1062
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure and function of a novel glycerophosphodiesterase from Enterobacter aerogenes.
|
|
C.J.Jackson,
P.D.Carr,
J.W.Liu,
S.J.Watt,
J.L.Beck,
D.L.Ollis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the glycerophosphodiesterase (GDPD) from Enterobacter
aerogenes, GpdQ, has been solved by SAD phasing from the active site metal ions.
Structural analysis indicates that GpdQ belongs to the alpha/beta sandwich
metallo-phosphoesterase family, rather than the (alpha/beta)(8) barrel GDPD
family, suggesting that GpdQ is a structurally novel GDPD. Hexameric GpdQ is
generated by interactions between three dimers. The dimers are formed through
domain swapping, stabilised by an inter-chain disulfide bond, and beta-sheet
extension. The active site contains a binuclear metal centre, with a fully
occupied alpha-metal ion site, and partially occupied beta-metal ion site, as
revealed by anomalous scattering analysis. Using a combination of TLS refinement
and normal mode analysis, the dynamic movement of GpdQ was investigated. This
analysis suggests that the hexameric quaternary structure stabilises the base of
the dimer, which promotes "breathing" of the active site cleft.
Comparison with other metallo-phosphodiesterases shows that although the
central, catalytic, domain is highly conserved, many of these enzymes possess
structurally unrelated secondary domains located at the entrance of the active
site. We suggest that this could be a common structural feature of
metallo-phosphodiesterases that constrains substrate specificity, preventing
non-specific phosphodiester hydrolysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The hydrolysis of the glycerophosphodiester
sn-glycero-3-phosphoethanolamine. The leaving group will be
protonated at physiological pH.
|
 |
Figure 2.
Figure 2. (Top) A topology diagram of GpdQ, illustrating the
catalytic α/β sandwich domain (β1–β7/αA–αF; 1–196),
the all β-strand dimerisation domain (β8–β12; 197–256),
and the domain swapped cap domain (β13, αG; 257–271) that is
stabilised by an inter-chain disulfide bond. The locations of
the liganding residues are shown. (Below) A ribbon diagram and
carbon-α trace (red) of dimeric GpdQ, showing the location of
the active site metals at the center of the α/β sandwich
domain and the disulfide bond (yellow).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
367,
1047-1062)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Foo,
C.J.Jackson,
P.D.Carr,
H.K.Kim,
G.Schenk,
L.R.Gahan,
and
D.L.Ollis
(2010).
Mutation of outer-shell residues modulates metal ion co-ordination strength in a metalloenzyme.
|
| |
Biochem J,
429,
313-321.
|
 |
|
|
|
|
 |
J.A.Larrabee,
W.R.Johnson,
and
A.S.Volwiler
(2009).
Magnetic circular dichroism study of a dicobalt(II) complex with mixed 5- and 6-coordination: a spectroscopic model for dicobalt(II) hydrolases.
|
| |
Inorg Chem,
48,
8822-8829.
|
 |
|
|
|
|
 |
M.Podobnik,
R.Tyagi,
N.Matange,
U.Dermol,
A.K.Gupta,
R.Mattoo,
K.Seshadri,
and
S.S.Visweswariah
(2009).
A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability.
|
| |
J Biol Chem,
284,
32846-32857.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.K.Dhar,
C.S.Thwin,
K.Tun,
Y.Tsumoto,
S.Maurer-Stroh,
F.Eisenhaber,
and
U.Surana
(2009).
Synthesizing non-natural parts from natural genomic template.
|
| |
J Biol Eng,
3,
2.
|
 |
|
|
|
|
 |
C.J.Jackson,
K.S.Hadler,
P.D.Carr,
A.J.Oakley,
S.Yip,
G.Schenk,
and
D.L.Ollis
(2008).
Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
681-685.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.S.Hadler,
T.Huber,
A.I.Cassady,
J.Weber,
J.Robinson,
A.Burrows,
G.Kelly,
L.W.Guddat,
D.A.Hume,
G.Schenk,
and
J.U.Flanagan
(2008).
Identification of a non-purple tartrate-resistant acid phosphatase: an evolutionary link to Ser/Thr protein phosphatases?
|
| |
BMC Res Notes,
1,
78.
|
 |
|
|
|
|
 |
L.Shi,
J.F.Liu,
X.M.An,
and
D.C.Liang
(2008).
Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism.
|
| |
Proteins,
72,
280-288.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.E.Mirams,
S.J.Smith,
K.S.Hadler,
D.L.Ollis,
G.Schenk,
and
L.R.Gahan
(2008).
Cadmium(II) complexes of the glycerophosphodiester-degrading enzyme GpdQ and a biomimetic N,O ligand.
|
| |
J Biol Inorg Chem,
13,
1065-1072.
|
 |
|
|
|
|
 |
S.Biswas,
R.J.Russell,
C.J.Jackson,
M.Vidovic,
O.Ganeshina,
J.G.Oakeshott,
and
C.Claudianos
(2008).
Bridging the synaptic gap: neuroligins and neurexin I in Apis mellifera.
|
| |
PLoS ONE,
3,
e3542.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

