 |
PDBsum entry 2df1
|
|

|
| Superseded by: |
 |
|
 |
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the extracellular domain of human cd38
|
|
Structure:
|
 |
Adp-ribosyl cyclase 1. Chain: a, b. Fragment: extracellular domain. Synonym: cyclic adp-ribose hydrolase 1, cadpr hydrolase 1, lymphocyte differentiation antigen cd38, t10, acute lymphoblastic leukemia cells antigen cd38. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Gene: cd38. Expressed in: escherichia coli.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.219
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
M.Kukimoto-Niino,O.Nureki,K.Murayama,M.Shirouzu,T.Katada, M.Hara-Yokoyama,S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
M.Kukimoto-Niino
et al.
Crystal structure of the extracellular domain of human cd38.
To be published,
.
|
 |
|
Date:
|
 |
|
22-Feb-06
|
Release date:
|
30-Jan-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P28907
(CD38_HUMAN) -
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
300 a.a.
247 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.2.5
- NAD(+) glycohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
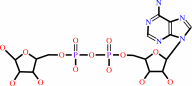 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

