 |
PDBsum entry 2dcf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.46
- 6-aminohexanoate-oligomer exohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
[N-(6-aminohexanoyl)](n) + H2O = [N-(6-aminohexanoyl)](n-1) + 6-aminohexanoate
|
|
2.
|
N-(6-aminohexanoyl)-6-aminohexanoate + H2O = 2 6-aminohexanoate
|
|
 |
 |
 |
 |
 |
[N-(6-aminohexanoyl)](n)
|
+
|
H2O
|
=
|
[N-(6-aminohexanoyl)](n-1)
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 6-aminohexanoate
6-aminohexanoate
|
|
 |
 |
 |
 |
 |
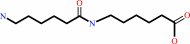 N-(6-aminohexanoyl)-6-aminohexanoate
N-(6-aminohexanoyl)-6-aminohexanoate
|
+
|
H2O
|
=
|
2
×
6-aminohexanoate
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
370:142-156
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase.
|
|
S.Negoro,
T.Ohki,
N.Shibata,
K.Sasa,
H.Hayashi,
H.Nakano,
K.Yasuhira,
D.Kato,
M.Takeo,
Y.Higuchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We performed X-ray crystallographic analyses of 6-aminohexanoate-dimer hydrolase
(Hyb-24DN), an enzyme responsible for the degradation of nylon-6, an industry
by-product, and of a complex between Hyb-24DN-A(112) (S112A-mutant of Hyb-24DN)
and 6-aminohexanoate-linear dimer (Ald) at 1.58 A and 1.4 A resolution,
respectively. In Hyb-24DN, Asp181-O(delta) forms hydrogen bonds with
Tyr170-O(eta), -two of the catalytic and binding amino acids, and a loop between
Asn167 and Val177. This state is the so-called open form, allowing its substrate
to bind in the space between the loop and catalytic residues. Upon substrate
binding (in Hyb-24DN-A(112)/Ald complex), the loop is shifted 4.3 A at
Tyr170-C(alpha), and the side-chain of Tyr170 is rotated. By the combined
effect, Tyr170-O(eta) moves a total of 10.5 A, resulting in the formation of
hydrogen bonds with the nitrogen of amide linkage in Ald (closed form). In
addition, electrostatic interaction between Asp181-O(delta) and the amino group
in Ald stabilizes the substrate binding. We propose here that the enzyme
catalysis proceeds according to the following steps: (i) Ald-induced transition
from open to closed form, (ii) nucleophilic attack of Ser112 to Ald and
formation of a tetrahedral intermediate, (iii) formation of acyl enzyme and
transition to open form, (iv) deacylation. Amino acid substitutions reducing the
enzyme/Ald interaction at positions 181 or 170 drastically decreased the
Ald-hydrolytic activity, but had very little effect on esterolytic activity,
suggesting that esterolytic reaction proceeds regardless of conversion. Present
models illustrate why new activity against the nylon oligomer has evolved in an
esterase with beta-lactamase folds, while retaining the original esterolytic
functions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Surface structure of entrance of catalytic
cleft of Hyb-24DN, DD-peptidase and class C β-lactamase.
(a) Hyb-24DN including Ald at spatially equivalent position
(open form). (b) Hyb-24DN-A^112/Ald complex (closed form). (c)
DD-Peptidase/substrate
(glycyl-L-α-amino-ε-pimelyl-D-alanyl-D-alanine) complex. (d)
Extended spectrum class C
β-lactamase/cefotaxime-analogue(m-nitrophenyl-2-(2-aminothiazol-4-yl)-2-[(Z)-methoxyimino]acetylaminomethyl
phosphonate) complex (PDB ID code: 1RGY). Substrates are shown
as stick model. Figures were generated with program MolFeat
(version 2.2, FiatLux Co.). Figure 5. Surface structure of
entrance of catalytic cleft of Hyb-24DN, DD-peptidase
and class C β-lactamase. (a) Hyb-24DN including Ald at
spatially equivalent position (open form). (b)
Hyb-24DN-A^112/Ald complex (closed form). (c)
DD-Peptidase/substrate
(glycyl-L-α-amino-ε-pimelyl-D-alanyl-D-alanine) complex. (d)
Extended spectrum class C
β-lactamase/cefotaxime-analogue(m-nitrophenyl-2-(2-aminothiazol-4-yl)-2-[(Z)-methoxyimino]acetylaminomethyl
phosphonate) complex (PDB ID code: 1RGY). Substrates are shown
as stick model. Figures were generated with program MolFeat
(version 2.2, FiatLux Co.).
|
 |
Figure 8.
Figure 8. Proposed catalytic mechanism of
6-aminohexanoate-dimer hydrolase. In this model, enzyme
catalysis proceeds according to the following steps: (i)
Ald-induced transition from open to closed form, (ii)
nucleophilic attack of Ser112 to Ald and formation of
tetrahedral intermediate, (iii) formation of acyl enzyme and
transition to open form, (iv) deacylation (formation of
tetrahedral intermediate and regeneration of free enzyme). (a)
Free enzyme, (d) acyl enzyme, and (e) tetrahedral intermediate
are present as open forms, and (b) enzyme + substrate and (c)
tetrahedral intermediate are present as closed forms. Figure
8. Proposed catalytic mechanism of 6-aminohexanoate-dimer
hydrolase. In this model, enzyme catalysis proceeds according to
the following steps: (i) Ald-induced transition from open to
closed form, (ii) nucleophilic attack of Ser112 to Ald and
formation of tetrahedral intermediate, (iii) formation of
acyl enzyme and transition to open form, (iv) deacylation
(formation of tetrahedral intermediate and regeneration of free
enzyme). (a) Free enzyme, (d) acyl enzyme, and (e) tetrahedral
intermediate are present as open forms, and (b) enzyme +
substrate and (c) tetrahedral intermediate are present as closed
forms.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
370,
142-156)
copyright 2007.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Nishizawa,
Y.Yabusaki,
and
M.Kanaoka
(2011).
Identification of the catalytic residues of carboxylesterase from Arthrobacter globiformis by diisopropyl fluorophosphate-labeling and site-directed mutagenesis.
|
| |
Biosci Biotechnol Biochem,
75,
89-94.
|
 |
|
|
|
|
 |
K.Yasuhira,
N.Shibata,
G.Mongami,
Y.Uedo,
Y.Atsumi,
Y.Kawashima,
A.Hibino,
Y.Tanaka,
Y.H.Lee,
D.Kato,
M.Takeo,
Y.Higuchi,
and
S.Negoro
(2010).
X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation.
|
| |
J Biol Chem,
285,
1239-1248.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Heumann,
A.Eberl,
G.Fischer-Colbrie,
H.Pobeheim,
F.Kaufmann,
D.Ribitsch,
A.Cavaco-Paulo,
and
G.M.Guebitz
(2009).
A novel aryl acylamidase from Nocardia farcinica hydrolyses polyamide.
|
| |
Biotechnol Bioeng,
102,
1003-1011.
|
 |
|
|
|
|
 |
T.Ohki,
N.Shibata,
Y.Higuchi,
Y.Kawashima,
M.Takeo,
D.Kato,
and
S.Negoro
(2009).
Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
|
| |
Protein Sci,
18,
1662-1673.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Kawashima,
T.Ohki,
N.Shibata,
Y.Higuchi,
Y.Wakitani,
Y.Matsuura,
Y.Nakata,
M.Takeo,
D.Kato,
and
S.Negoro
(2009).
Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
|
| |
FEBS J,
276,
2547-2556.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.M.Guebitz,
and
A.Cavaco-Paulo
(2008).
Enzymes go big: surface hydrolysis and functionalization of synthetic polymers.
|
| |
Trends Biotechnol,
26,
32-38.
|
 |
|
|
|
|
 |
S.Okazaki,
A.Suzuki,
H.Komeda,
Y.Asano,
and
T.Yamane
(2008).
Deduced catalytic mechanism of D-amino acid amidase from Ochrobactrum anthropi SV3.
|
| |
J Synchrotron Radiat,
15,
250-253.
|
 |
|
|
|
|
 |
K.Yasuhira,
Y.Tanaka,
H.Shibata,
Y.Kawashima,
A.Ohara,
D.Kato,
M.Takeo,
and
S.Negoro
(2007).
6-Aminohexanoate oligomer hydrolases from the alkalophilic bacteria Agromyces sp. strain KY5R and Kocuria sp. strain KY2.
|
| |
Appl Environ Microbiol,
73,
7099-7102.
|
 |
|
|
|
|
 |
K.Yasuhira,
Y.Uedo,
M.Takeo,
D.Kato,
and
S.Negoro
(2007).
Genetic organization of nylon-oligomer-degrading enzymes from alkalophilic bacterium, Agromyces sp. KY5R.
|
| |
J Biosci Bioeng,
104,
521-524.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

