 |
PDBsum entry 2cvs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2cvs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
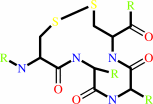 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
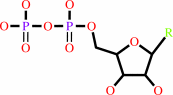 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
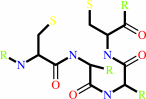 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
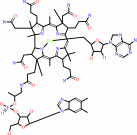 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:4022-4027
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation.
|
|
H.Xu,
C.Faber,
T.Uchiki,
J.W.Fairman,
J.Racca,
C.Dealwis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ribonucleotide reductase catalyzes a crucial step in de novo DNA synthesis and
is allosterically controlled by relative levels of dNTPs to maintain a balanced
pool of deoxynucleoside triphosphates in the cell. In eukaryotes, the enzyme
comprises a heterooligomer of alpha(2) and beta(2) subunits. The alpha subunit,
Rnr1, contains catalytic and regulatory sites. Here, we report the only x-ray
structures of the eukaryotic alpha subunit of ribonucleotide reductase from
Saccharomyces cerevisiae. The structures of the apo-, AMPPNP only-, AMPPNP-CDP-,
AMPPNP-UDP-, dGTP-ADP- and TTP-GDP-bound complexes give insight into substrate
and effector binding and specificity cross-talk. These are Class I structures
with the only fully ordered catalytic sites, including loop 2, a stretch of
polypeptide that spans specificity and catalytic sites, conferring specificity.
Binding of specificity effector rearranges loop 2; in our structures, this
rearrangement moves P294, a residue unique to eukaryotes, out of the catalytic
site, accommodating substrate binding. Substrate binding further rearranges loop
2. Cross-talk, by which effector binding regulates substrate preference, occurs
largely through R293 and Q288 of loop 2, which are analogous to residues in
Thermotoga maritima that mediate cross-talk. However loop-2 conformations and
residue-substrate interactions differ substantially between yeast and T.
maritima. In most effector-substrate complexes, water molecules help mediate
substrate-loop 2 interactions. Finally, the substrate ribose binds with its 3'
hydroxyl closer than its 2' hydroxyl to C218 of the catalytic redox pair. We
also see a conserved water molecule at the catalytic site in all our structures,
near the ribose 2' hydroxyl.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Structure of Rnr1. (A) The dimer of apo Rnr1. Rnr1
monomers are yellow and green; dGTP (violet) and ADP (blue) from
the cognate complex are shown in the specificity and catalytic
sites. The three-helix insert and C-terminal insert are red and
blue, respectively. (B) 1.0  2F[o]–F[c] electron
density for effector (blue density), loop 2 (red density), and
substrate (green density) for the dGTP–ADP complex. 2F[o]–F[c] electron
density for effector (blue density), loop 2 (red density), and
substrate (green density) for the dGTP–ADP complex.
|
 |
Figure 3.
Fig. 3. Specificity-site interactions. (A) Loop-2
rearrangements. Substrate (Left) and loop 2 and effector (Right)
are shown for AMPPNP–UDP (yellow). Loop 2 is shown for apo
(black), AMPPNP only (gray), and AMPPNP–CDP (orange). P294 is
shown for apo and AMPPNP only. Q288 and R293 are shown for
AMPPNP–CDP and AMPPNP–UDP. C  spheres are shown for
Q288, R293, and P294 in all structures. (B–E) Specificity
effector binding. Colors of secondary structure cartoons are as
in Fig. 2. Loop-1 carbons are yellow; Loop-2 carbons, light
blue; other interacting atoms are colored as in Fig. 2, except
effector carbons, which are green. (B) Stereoview of dGTP–ADP.
(C) Stereoview of TTP–GDP. (D) Stereoview of AMPPNP–CDP. (E)
Stereoview of AMPPNP–UDP. spheres are shown for
Q288, R293, and P294 in all structures. (B–E) Specificity
effector binding. Colors of secondary structure cartoons are as
in Fig. 2. Loop-1 carbons are yellow; Loop-2 carbons, light
blue; other interacting atoms are colored as in Fig. 2, except
effector carbons, which are green. (B) Stereoview of dGTP–ADP.
(C) Stereoview of TTP–GDP. (D) Stereoview of AMPPNP–CDP. (E)
Stereoview of AMPPNP–UDP.
|
 |
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
First eukaryotic ribonucleotide reductase I structure explains dNTP
regulation.
Chris Dealwis
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Kumar,
A.L.Abdulovic,
J.Viberg,
A.K.Nilsson,
T.A.Kunkel,
and
A.Chabes
(2011).
Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools.
|
| |
Nucleic Acids Res,
39,
1360-1371.
|
 |
|
|
|
|
 |
D.T.Logan
(2011).
Closing the circle on ribonucleotide reductases.
|
| |
Nat Struct Mol Biol,
18,
251-253.
|
 |
|
|
|
|
 |
A.Holmgren,
and
R.Sengupta
(2010).
The use of thiols by ribonucleotide reductase.
|
| |
Free Radic Biol Med,
49,
1617-1628.
|
 |
|
|
|
|
 |
D.Kumar,
J.Viberg,
A.K.Nilsson,
and
A.Chabes
(2010).
Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint.
|
| |
Nucleic Acids Res,
38,
3975-3983.
|
 |
|
|
|
|
 |
J.J.Kohler,
S.H.Hosseini,
I.Cucoranu,
O.Zhelyabovska,
E.Green,
K.Ivey,
A.Abuin,
E.Fields,
A.Hoying,
R.Russ,
R.Santoianni,
C.M.Raper,
Q.Yang,
A.Lavie,
and
W.Lewis
(2010).
Transgenic cardiac-targeted overexpression of human thymidylate kinase.
|
| |
Lab Invest,
90,
383-390.
|
 |
|
|
|
|
 |
D.Sun,
H.Xu,
S.R.Wijerathna,
C.Dealwis,
and
R.E.Lee
(2009).
Structure-Based Design, Synthesis, and Evaluation of 2'-(2-Hydroxyethyl)-2'-deoxyadenosine and the 5'-Diphosphate Derivative as Ribonucleotide Reductase Inhibitors.
|
| |
ChemMedChem,
4,
1649-1656.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Artin,
J.Wang,
G.J.Lohman,
K.Yokoyama,
G.Yu,
R.G.Griffin,
G.Bar,
and
J.Stubbe
(2009).
Insight into the mechanism of inactivation of ribonucleotide reductase by gemcitabine 5'-diphosphate in the presence or absence of reductant.
|
| |
Biochemistry,
48,
11622-11629.
|
 |
|
|
|
|
 |
T.Radivoyevitch
(2009).
Automated mass action model space generation and analysis methods for two-reactant combinatorially complex equilibriums: an analysis of ATP-induced ribonucleotide reductase R1 hexamerization data.
|
| |
Biol Direct,
4,
50.
|
 |
|
|
|
|
 |
H.Xu,
J.W.Fairman,
S.R.Wijerathna,
N.R.Kreischer,
J.LaMacchia,
E.Helmbrecht,
B.S.Cooperman,
and
C.Dealwis
(2008).
The structural basis for peptidomimetic inhibition of eukaryotic ribonucleotide reductase: a conformationally flexible pharmacophore.
|
| |
J Med Chem,
51,
4653-4659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.H.Kim
(2007).
Outliers in SAR and QSAR: 2. Is a flexible binding site a possible source of outliers?
|
| |
J Comput Aided Mol Des,
21,
421-435.
|
 |
|
|
|
|
 |
Z.Zhang,
K.Yang,
C.C.Chen,
J.Feser,
and
M.Huang
(2007).
Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1.
|
| |
Proc Natl Acad Sci U S A,
104,
2217-2222.
|
 |
|
|
|
|
 |
H.Xu,
C.Faber,
T.Uchiki,
J.Racca,
and
C.Dealwis
(2006).
Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly.
|
| |
Proc Natl Acad Sci U S A,
103,
4028-4033.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

