 |
PDBsum entry 2c29

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2c29
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.219
- dihydroflavanol 4-reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Flavonoid Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (2R,3S,4S)-leucoanthocyanidin + NADP+ = a (2R,3R)-dihydroflavonol + NADPH + H+
|
 |
 |
 |
 |
 |
(2R,3S,4S)-leucoanthocyanidin
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
(2R,3R)-dihydroflavonol
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.234
- flavanone 4-reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2S)-flavan-4-ol + NADP+ = (2S)-flavanone + NADPH + H+
|
 |
 |
 |
 |
 |
(2S)-flavan-4-ol
Bound ligand (Het Group name = )
matches with 77.27% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
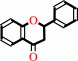 (2S)-flavanone
(2S)-flavanone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
368:1345-1357
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis.
|
|
P.Petit,
T.Granier,
B.L.d'Estaintot,
C.Manigand,
K.Bathany,
J.M.Schmitter,
V.Lauvergeat,
S.Hamdi,
B.Gallois.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The nicotinamide adenine dinucleotide phosphate (NADPH)-dependent enzyme
dihydroflavonol 4-reductase (DFR) catalyzes a late step in the biosynthesis of
anthocyanins and condensed tannins, two flavonoid classes of importance to plant
survival and human nutrition. This enzyme has been widely investigated in many
plant species, but little is known about its structural and biochemical
properties. To provide a basis for detailed structure-function studies, the
crystal structure of Vitis vinifera DFR, heterologously expressed in Escherichia
coli, has been determined at 1.8 A resolution. The 3D structure of the ternary
complex obtained with the oxidized form of nicotinamide adenine dinucleotide
phosphate and dihydroquercetin, one of the DFR substrates, presents common
features with the short-chain dehydrogenase/reductase family, i.e., an
N-terminal domain adopting a Rossmann fold and a variable C-terminal domain,
which participates in substrate binding. The structure confirms the importance
of the 131-156 region, which lines the substrate binding site and enlightens the
role of a specific residue at position 133 (Asn or Asp), assumed to control
substrate recognition. The activity of the wild-type enzyme and its variant
N133D has been quantified in vitro, using dihydroquercetin or dihydrokaempferol.
Our results demonstrate that position 133 cannot be solely responsible for the
recognition of the B-ring hydroxylation pattern of dihydroflavonols.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Final steps of the flavonoid biosynthetic pathway
leading to the synthesis of anthocyanins and condensed tannins
(proanthocyanidins). DFR, dihydroflavonol 4-reductase; ANR,
anthocyanidin reductase; ANS, anthocyanidin synthase; LAR,
leucoanthocyanidin reductase; UFGT, 3-O-glycosyltransferase.
Figure 1. Final steps of the flavonoid biosynthetic pathway
leading to the synthesis of anthocyanins and condensed tannins
(proanthocyanidins). DFR, dihydroflavonol 4-reductase; ANR,
anthocyanidin reductase; ANS, anthocyanidin synthase; LAR,
leucoanthocyanidin reductase; UFGT, 3-O-glycosyltransferase.
|
 |
Figure 5.
Figure 5. (a) Geometry of the catalytic triad site. (b) Stereo
view of the catalytic site in the vicinity of the K167
side-chain. The lysine amino group interacts with a cluster of
five water molecules filling a hydrophilic cavity. (c) Stereo
view of the substrate binding site. DHQ is wrapped up by the N
and C-terminal parts of the protein: contacts are established
via hydrophilic interactions or hydrophobic residues of both N
and C-terminal domains. The color code is the same as in the
legend to Figure 4. Figure 5. (a) Geometry of the catalytic
triad site. (b) Stereo view of the catalytic site in the
vicinity of the K167 side-chain. The lysine amino group
interacts with a cluster of five water molecules filling a
hydrophilic cavity. (c) Stereo view of the substrate binding
site. DHQ is wrapped up by the N and C-terminal parts of the
protein: contacts are established via hydrophilic interactions
or hydrophobic residues of both N and C-terminal domains. The
color code is the same as in the legend to [3]Figure 4.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
368,
1345-1357)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Wang
(2011).
Structure, function, and engineering of enzymes in isoflavonoid biosynthesis.
|
| |
Funct Integr Genomics,
11,
13-22.
|
 |
|
|
|
|
 |
H.Halbwirth
(2010).
The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway.
|
| |
Int J Mol Sci,
11,
595-621.
|
 |
|
|
|
|
 |
M.Gargouri,
J.Chaudière,
C.Manigand,
C.Maugé,
K.Bathany,
J.M.Schmitter,
and
B.Gallois
(2010).
The epimerase activity of anthocyanidin reductase from Vitis vinifera and its regiospecific hydride transfers.
|
| |
Biol Chem,
391,
219-227.
|
 |
|
|
|
|
 |
M.Katzberg,
N.Skorupa-Parachin,
M.F.Gorwa-Grauslund,
and
M.Bertau
(2010).
Engineering cofactor preference of ketone reducing biocatalysts: A mutagenesis study on a gamma-diketone reductase from the yeast Saccharomyces cerevisiae serving as an example.
|
| |
Int J Mol Sci,
11,
1735-1758.
|
 |
|
|
|
|
 |
M.Gargouri,
C.Manigand,
C.Maugé,
T.Granier,
B.Langlois d'Estaintot,
O.Cala,
I.Pianet,
K.Bathany,
J.Chaudière,
and
B.Gallois
(2009).
Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
989.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhang,
S.Cheng,
D.De Jong,
H.Griffiths,
R.Halitschke,
and
W.De Jong
(2009).
The potato R locus codes for dihydroflavonol 4-reductase.
|
| |
Theor Appl Genet,
119,
931-937.
|
 |
|
|
|
|
 |
D.L.Des Marais,
and
M.D.Rausher
(2008).
Escape from adaptive conflict after duplication in an anthocyanin pathway gene.
|
| |
Nature,
454,
762-765.
|
 |
|
|
|
|
 |
N.Trabelsi,
P.Petit,
C.Manigand,
B.Langlois d'Estaintot,
T.Granier,
J.Chaudière,
and
B.Gallois
(2008).
Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols.
|
| |
Acta Crystallogr D Biol Crystallogr,
(),
883-891.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

