 |
PDBsum entry 2c0r

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.52
- phosphoserine transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
O-phospho-L-serine + 2-oxoglutarate = 3-phosphooxypyruvate + L-glutamate
|
|
2.
|
4-(phosphooxy)-L-threonine + 2-oxoglutarate = (R)-3-hydroxy-2-oxo-4- phosphooxybutanoate + L-glutamate
|
|
 |
 |
 |
 |
 |
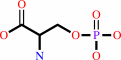 O-phospho-L-serine
O-phospho-L-serine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
3-phosphooxypyruvate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
4-(phosphooxy)-L-threonine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
(R)-3-hydroxy-2-oxo-4- phosphooxybutanoate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
63:742-753
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Effect of pH on the structure and stability of Bacillus circulans ssp. alkalophilus phosphoserine aminotransferase: thermodynamic and crystallographic studies.
|
|
E.G.Kapetaniou,
A.Thanassoulas,
A.P.Dubnovitsky,
G.Nounesis,
A.C.Papageorgiou.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
pH is one of the key parameters that affect the stability and function of
proteins. We have studied the effect of pH on the
pyridoxal-5'-phosphate-dependent enzyme phosphoserine aminotransferase produced
by the facultative alkaliphile Bacillus circulans ssp. alkalophilus using
thermodynamic and crystallographic analysis. Enzymatic activity assay showed
that the enzyme has maximum activity at pH 9.0 and relative activity less than
10% at pH 7.0. Differential scanning calorimetry and circular dichroism
experiments revealed variations in the stability and denaturation profiles of
the enzyme at different pHs. Most importantly, release of pyridoxal-5'-phosphate
and protein thermal denaturation were found to occur simultaneously at pH 6.0 in
contrast to pH 8.5 where denaturation preceded cofactor's release by
approximately 3 degrees C. To correlate the observed differences in thermal
denaturation with structural features, the crystal structure of phosphoserine
aminotransferase was determined at 1.2 and 1.5 A resolution at two different pHs
(8.5 and 4.6, respectively). Analysis of the two structures revealed changes in
the vicinity of the active site and in surface residues. A conformational change
in a loop involved in substrate binding at the entrance of the active site has
been identified upon pH change. Moreover, the number of intramolecular ion pairs
was found reduced in the pH 4.6 structure. Taken together, the presented
kinetics, thermal denaturation, and crystallographic data demonstrate a
potential role of the active site in unfolding and suggest that subtle but
structurally significant conformational rearrangements are involved in the
stability and integrity of phosphoserine aminotransferase in response to pH
changes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Schematic representation of the chemical reaction
catalyzed by phosphoserine aminotransferase.
|
 |
Figure 7.
Figure 7. Ribbon representation of the BCIR PSAT structure at
pH 8.5 (A) dimer (B) monomer. Secondary-structure elements were
calculated using DSSP.[29] The  -helices
are shown in yellow and the -helices
are shown in yellow and the  -strands
in cyan. N- and C-termini are shown as spheres and PLP molecules
in space filling (A) and ball-and-stick (B). The small domain
contains an extended five-stranded -strands
in cyan. N- and C-termini are shown as spheres and PLP molecules
in space filling (A) and ball-and-stick (B). The small domain
contains an extended five-stranded  -sheet,
composed of a two-stranded parallel -sheet,
composed of a two-stranded parallel  -sheet
( -sheet
(  1
and 1
and  13)
and a three-stranded antiparallel 13)
and a three-stranded antiparallel  -sheet
(+ -sheet
(+  11,
- 11,
-  12,
- 12,
-  14).
The large domain contains the conservative among PLP-dependent
enzymes[44] seven-stranded 14).
The large domain contains the conservative among PLP-dependent
enzymes[44] seven-stranded  -sheet
( -sheet
(  2- 2-
 5
and 5
and  7- 7-
 9)
with all 9)
with all  -strands
parallel except -strands
parallel except  9.
Two additional 9.
Two additional  -strands
( -strands
(  6
and 6
and  10),
six 10),
six  -helices
( -helices
(  1- 1-
 6),
and six 3[10]-helices complete the large domain. Helix 6),
and six 3[10]-helices complete the large domain. Helix  7
connects the large domain with the small domain. The
five-stranded 7
connects the large domain with the small domain. The
five-stranded  -sheet
is flanked by helix -sheet
is flanked by helix  7
and two additional helices ( 7
and two additional helices (  8
and 8
and  9)
close to the C-terminus. 9)
close to the C-terminus.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2006,
63,
742-753)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
V.Mishra,
V.Ali,
T.Nozaki,
and
V.Bhakuni
(2011).
Biophysical characterization of Entamoeba histolytica phosphoserine aminotransferase (EhPSAT): role of cofactor and domains in stability and subunit assembly.
|
| |
Eur Biophys J,
40,
599-610.
|
 |
|
|
|
|
 |
V.Mishra,
V.Ali,
T.Nozaki,
and
V.Bhakuni
(2010).
Entamoeba histolytica Phosphoserine aminotransferase (EhPSAT): insights into the structure-function relationship.
|
| |
BMC Res Notes,
3,
52.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

