 |
PDBsum entry 2bf4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Reductase
|
 |
|
Title:
|
 |
A second fmn-binding site in yeast NADPH-cytochrome p450 reductase suggests a novel mechanism of electron transfer by diflavin reductases.
|
|
Structure:
|
 |
NADPH-cytochrome p450 reductase. Chain: a, b. Synonym: cpr, p450r. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.197
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
L.M.Podust,G.I.Lepesheva,Y.Kim,L.V.Yermalitskaya,V.N.Yermalitsky, D.C.Lamb,S.L.Kelly,M.R.Waterman
|
Key ref:

|
 |
D.C.Lamb
et al.
(2006).
A second FMN binding site in yeast NADPH-cytochrome P450 reductase suggests a mechanism of electron transfer by diflavin reductases.
Structure,
14,
51-61.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
03-Dec-04
|
Release date:
|
17-Jan-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P16603
(NCPR_YEAST) -
NADPH--cytochrome P450 reductase from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
691 a.a.
645 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.6.2.4
- NADPH--hemoprotein reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 oxidized [cytochrome P450] + NADPH = 2 reduced [cytochrome P450] + NADP+ + H+
|
 |
 |
 |
 |
 |
2
×
oxidized [cytochrome P450]
|
+
|
 NADPH
NADPH
|
=
|
2
×
reduced [cytochrome P450]
|
+
|
 NADP(+)
NADP(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
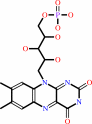 FMN
FMN
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
14:51-61
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
A second FMN binding site in yeast NADPH-cytochrome P450 reductase suggests a mechanism of electron transfer by diflavin reductases.
|
|
D.C.Lamb,
Y.Kim,
L.V.Yermalitskaya,
V.N.Yermalitsky,
G.I.Lepesheva,
S.L.Kelly,
M.R.Waterman,
L.M.Podust.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
NADPH-cytochrome P450 reductase transfers two reducing equivalents derived from
a hydride ion of NADPH via FAD and FMN to the large family of microsomal
cytochrome P450 monooxygenases in one-electron transfer steps. The mechanism of
electron transfer by diflavin reductases remains elusive and controversial.
Here, we determined the crystal structure of truncated yeast NADPH-cytochrome
P450 reductase, which is functionally active toward its physiological substrate
cytochrome P450, and discovered a second FMN binding site at the interface of
the connecting and FMN binding domains. The two FMN binding sites have different
accessibilities to the bulk solvent and different amino acid environments,
suggesting stabilization of different electronic structures of the reduced
flavin. Since only one FMN cofactor is required for function, a hypothetical
mechanism of electron transfer is discussed that proposes shuttling of a single
FMN between these two sites coupled with the transition between two semiquinone
forms, neutral (blue) and anionic (red).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Hypothetical Mechanism of Electron Transfer by
P450 Reductases
(A) Single catalytic turnover of yCPR.
Neutral semiquinone is shown in blue, and putative anionic
semiquinone is shown in red.
(B) Structures of oxidized and
semiquinone states of the isoalloxazine system shown with the si
side facing the viewer.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2006,
14,
51-61)
copyright 2006.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Aigrain,
D.Pompon,
and
G.Truan
(2011).
Role of the interface between the FMN and FAD domains in the control of redox potential and electronic transfer of NADPH-cytochrome P450 reductase.
|
| |
Biochem J,
435,
197-206.
|
 |
|
|
|
|
 |
A.Das,
and
S.G.Sligar
(2009).
Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer.
|
| |
Biochemistry,
48,
12104-12112.
|
 |
|
|
|
|
 |
D.Hamdane,
C.Xia,
S.C.Im,
H.Zhang,
J.J.Kim,
and
L.Waskell
(2009).
Structure and Function of an NADPH-Cytochrome P450 Oxidoreductase in an Open Conformation Capable of Reducing Cytochrome P450.
|
| |
J Biol Chem,
284,
11374-11384.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Aigrain,
D.Pompon,
G.Truan,
and
S.Moréra
(2009).
Cloning, purification, crystallization and preliminary X-ray analysis of a chimeric NADPH-cytochrome P450 reductase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
210-212.
|
 |
|
|
|
|
 |
L.Aigrain,
D.Pompon,
S.Moréra,
and
G.Truan
(2009).
Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase.
|
| |
EMBO Rep,
10,
742-747.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Mendieta-Wejebe,
M.C.Rosales-Hernández,
H.Rios,
J.Trujillo-Ferrara,
G.López-Pérez,
F.Tamay-Cach,
R.Ramos-Morales,
and
J.Correa-Basurto
(2008).
Comparing the electronic properties and docking calculations of heme derivatives on CYP2B4.
|
| |
J Mol Model,
14,
537-545.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

