 |
PDBsum entry 2anh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Alkaline phosphatase
|
PDB id
|
|

|

|
2anh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.1
- alkaline phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phosphate monoester + H2O = an alcohol + phosphate
|
 |
 |
 |
 |
 |
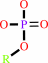 phosphate monoester
phosphate monoester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+); Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
253:604-617
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
|
|
J.E.Murphy,
T.T.Tibbitts,
E.R.Kantrowitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In order to understand some of the differences between human placental, human,
Saccharomyces cerevisiae and Escherichia coli alkaline phosphatases in specific
activity, activation by magnesium, and pH versus activity profiles, the X-ray
crystal structures of three mutant E. coli alkaline phosphatases have been
determined. The aligned sequences of alkaline phosphatases from mammalian, yeast
and E. coli show that 25 to 30% of the amino acids are absolutely conserved and
the active site residues are completely conserved with the exception of residues
153, 328 and 155. The bacterial enzyme has a salt-bridge, Asp153/Lys328, near
the third metal binding site which, based on sequence homology, is apparently
absent in the yeast and mammalian enzymes. The human enzymes have histidine at
positions 153 and 328, and the yeast enzyme has histidine at position 328. In
the E. coli enzyme, Asp153 was replaced by histidine (D153H), Lys328 was
replaced by histidine (K328H), and a double mutant (DM) was constructed
containing both mutations. The structure of the K328H enzyme was refined using
cross-validation to a resolution of 2.3 A with a working R-factor of 0.181 and a
free R-factor of 0.249. The DM structure was determined to a resolution of 2.5 A
with a working R-factor of 0.166 and a free R-factor of 0.233. The structure of
the D135H enzyme, which has been reported to a resolution of 2.4 A, has been
re-refined using cross-validation to a working R-factor of 0.179 and a free
R-factor of 0.239 for controlled comparisons with the two new structures. In all
three structures the most significant changes are related to the bound phosphate
inhibitor and the identity of the metal ion in the third binding site. The
changes in the position of the phosphate group and the alterations at the third
metal binding site indicate the structural basis for the variations in the
steady-state kinetic parameters previously reported for these enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. The active site region of E. coli alkaline
phosphatase including the phosphate group, magnesium
ion and two zinc-binding sites. Not all the ligands are
shown. Water molecules are indicated by the letter w.
Hydrogen bonds are shown as broken lines (Kim &
Wyckoff, 1991).
|
 |
Figure 9.
Figure 9. Stereoview comparing the wild-type (thinnest lines), the K328H (middle lines) and the DM (thickest lines)
structures. This view focuses on the new anion binding site (PO4-B) around Tyr169. This site exists in the D153H and
DM structures, but is not seen in the K328H or the wild-type structure. The Figure shows the loop region formed by
the disulfide bridge between Cys168 and Cys178. The conformation of this loop region appears to be conserved in all
three structures.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1995,
253,
604-617)
copyright 1995.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Koutsioulis,
A.Lyskowski,
S.Mäki,
E.Guthrie,
G.Feller,
V.Bouriotis,
and
P.Heikinheimo
(2010).
Coordination sphere of the third metal site is essential to the activity and metal selectivity of alkaline phosphatases.
|
| |
Protein Sci,
19,
75-84.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Qiao,
C.Ellis,
J.Steffen,
C.Y.Wu,
and
D.J.Eide
(2009).
Zinc status and vacuolar zinc transporters control alkaline phosphatase accumulation and activity in Saccharomyces cerevisiae.
|
| |
Mol Microbiol,
72,
320-334.
|
 |
|
|
|
|
 |
Y.Zhang,
C.Ji,
X.Zhang,
Z.Yang,
J.Peng,
R.Qiu,
Y.Xie,
and
Y.Mao
(2008).
A moderately thermostable alkaline phosphatase from Geobacillus thermodenitrificans T2: cloning, expression and biochemical characterization.
|
| |
Appl Biochem Biotechnol,
151,
81-92.
|
 |
|
|
|
|
 |
J.Li,
L.Xu,
and
F.Yang
(2007).
Expression and characterization of recombinant thermostable alkaline phosphatase from a novel thermophilic bacterium Thermus thermophilus XM.
|
| |
Acta Biochim Biophys Sin (Shanghai),
39,
844-850.
|
 |
|
|
|
|
 |
R.Mitra,
M.W.Peters,
and
M.J.Scott
(2007).
Synthesis and reactivity of a C3-symmetric trinuclear zinc(II) hydroxide catalyst efficient at phosphate diester transesterification.
|
| |
Dalton Trans,
(),
3924-3935.
|
 |
|
|
|
|
 |
T.Harada,
I.Koyama,
T.Matsunaga,
A.Kikuno,
T.Kasahara,
M.Hassimoto,
D.H.Alpers,
and
T.Komoda
(2005).
Characterization of structural and catalytic differences in rat intestinal alkaline phosphatase isozymes.
|
| |
FEBS J,
272,
2477-2486.
|
 |
|
|
|
|
 |
M.M.de Backer,
S.McSweeney,
P.F.Lindley,
and
E.Hough
(2004).
Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1555-1561.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Kozlenkov,
T.Manes,
M.F.Hoylaerts,
and
J.L.Millán
(2002).
Function assignment to conserved residues in mammalian alkaline phosphatases.
|
| |
J Biol Chem,
277,
22992-22999.
|
 |
|
|
|
|
 |
C.L.Wojciechowski,
and
E.R.Kantrowitz
(2002).
Altering of the metal specificity of Escherichia coli alkaline phosphatase.
|
| |
J Biol Chem,
277,
50476-50481.
|
 |
|
|
|
|
 |
P.J.O'Brien,
and
D.Herschlag
(2002).
Alkaline phosphatase revisited: hydrolysis of alkyl phosphates.
|
| |
Biochemistry,
41,
3207-3225.
|
 |
|
|
|
|
 |
H.C.Hung,
and
G.G.Chang
(2001).
Differentiation of the slow-binding mechanism for magnesium ion activation and zinc ion inhibition of human placental alkaline phosphatase.
|
| |
Protein Sci,
10,
34-45.
|
 |
|
|
|
|
 |
H.C.Hung,
and
G.G.Chang
(2001).
Multiple unfolding intermediates of human placental alkaline phosphatase in equilibrium urea denaturation.
|
| |
Biophys J,
81,
3456-3471.
|
 |
|
|
|
|
 |
I.O.Fritsky,
R.Ott,
H.Pritzkow,
and
R.Krämer
(2001).
An allosteric synthetic catalyst: metal ions tune the activity of an artificial phosphodiesterase.
|
| |
Chemistry,
7,
1221-1231.
|
 |
|
|
|
|
 |
I.Tsigos,
K.Mavromatis,
M.Tzanodaskalaki,
C.Pozidis,
M.Kokkinidis,
and
V.Bouriotis
(2001).
Engineering the properties of a cold active enzyme through rational redesign of the active site.
|
| |
Eur J Biochem,
268,
5074-5080.
|
 |
|
|
|
|
 |
R.Q.Zhang,
Q.X.Chen,
R.Xiao,
L.P.Xie,
X.G.Zeng,
and
H.M.Zhou
(2001).
Inhibition kinetics of green crab (Scylla serrata) alkaline phosphatase by zinc ions: a new type of complexing inhibition.
|
| |
Biochim Biophys Acta,
1545,
6.
|
 |
|
|
|
|
 |
S.Zappa,
J.L.Rolland,
D.Flament,
Y.Gueguen,
J.Boudrant,
and
J.Dietrich
(2001).
Characterization of a highly thermostable alkaline phosphatase from the euryarchaeon Pyrococcus abyssi.
|
| |
Appl Environ Microbiol,
67,
4504-4511.
|
 |
|
|
|
|
 |
Y.D.Park,
Y.Yang,
Q.X.Chen,
H.N.Lin,
Q.Liu,
and
H.M.Zhou
(2001).
Kinetics of complexing activation by the magnesium ion on green crab (Scylla serrata) alkaline phosphatase.
|
| |
Biochem Cell Biol,
79,
765-772.
|
 |
|
|
|
|
 |
B.Asgeirsson,
J.B.Hauksson,
and
G.H.Gunnarsson
(2000).
Dissociation and unfolding of cold-active alkaline phosphatase from atlantic cod in the presence of guanidinium chloride.
|
| |
Eur J Biochem,
267,
6403-6412.
|
 |
|
|
|
|
 |
I.O.Fritsky,
R.Ott,
and
R.Krämer
(2000).
Allosteric Regulation of Artificial Phosphoesterase Activity by Metal Ions This work was funded by the DFG (Gerhard Hess Programm).
|
| |
Angew Chem Int Ed Engl,
39,
3255-3258.
|
 |
|
|
|
|
 |
Q.X.Chen,
W.Z.Zheng,
J.Y.Lin,
Y.Shi,
W.Z.Xie,
and
H.M.Zhou
(2000).
Effect of metal ions on the activity of green crab (Scylla serrata) alkaline phosphatase.
|
| |
Int J Biochem Cell Biol,
32,
879-885.
|
 |
|
|
|
|
 |
K.M.Holtz,
B.Stec,
and
E.R.Kantrowitz
(1999).
A model of the transition state in the alkaline phosphatase reaction.
|
| |
J Biol Chem,
274,
8351-8354.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Sun,
D.C.Martin,
and
E.R.Kantrowitz
(1999).
Rate-determining step of Escherichia coli alkaline phosphatase altered by the removal of a positive charge at the active center.
|
| |
Biochemistry,
38,
2842-2848.
|
 |
|
|
|
|
 |
M.Y.Galperin,
A.Bairoch,
and
E.V.Koonin
(1998).
A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases.
|
| |
Protein Sci,
7,
1829-1835.
|
 |
|
|
|
|
 |
T.Manes,
M.F.Hoylaerts,
R.Müller,
F.Lottspeich,
W.Hölke,
and
J.L.Millán
(1998).
Genetic complexity, structure, and characterization of highly active bovine intestinal alkaline phosphatases.
|
| |
J Biol Chem,
273,
23353-23360.
|
 |
|
|
|
|
 |
J.E.Murphy,
B.Stec,
L.Ma,
and
E.R.Kantrowitz
(1997).
Trapping and visualization of a covalent enzyme-phosphate intermediate.
|
| |
Nat Struct Biol,
4,
618-622.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

