 |
PDBsum entry 2a8y

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of 5'-deoxy-5'methylthioadenosine phosphorylase complexed with 5'-deoxy-5'methylthioadenosine and sulfate
|
|
Structure:
|
 |
5'-methylthioadenosine phosphorylase (mtap). Chain: a, b, c, d, e, f, g, h, i, j, k, l. Engineered: yes
|
|
Source:
|
 |
Sulfolobus solfataricus. Organism_taxid: 2287. Gene: ssmtapii. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Hexamer (from
)
Hexamer (from
)
|
|
Resolution:
|
 |
|
1.45Å
|
R-factor:
|
0.182
|
R-free:
|
0.202
|
|
|
Authors:
|
 |
Y.Zhang,M.Porcelli,G.Cacciapuoti,S.E.Ealick
|
Key ref:

|
 |
Y.Zhang
et al.
(2006).
The crystal structure of 5'-deoxy-5'-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds.
J Mol Biol,
357,
252-262.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Jul-05
|
Release date:
|
28-Mar-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q97W94
(MTAP_SULSO) -
S-methyl-5'-thioadenosine phosphorylase from Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
270 a.a.
263 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.28
- S-methyl-5'-thioadenosine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-methyl-5'-thioadenosine + phosphate = 5-(methylsulfanyl)-alpha-D-ribose 1-phosphate + adenine
|
 |
 |
 |
 |
 |
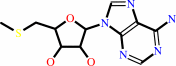 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
5-(methylsulfanyl)-alpha-D-ribose 1-phosphate
|
+
|
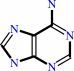 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
357:252-262
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of 5'-deoxy-5'-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds.
|
|
Y.Zhang,
M.Porcelli,
G.Cacciapuoti,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of Sulfolobus solfataricus 5'-deoxy-5'-methylthioadenosine
phosphorylase II (SsMTAPII) in complex with 5'-deoxy-5'-methylthioadenosine
(MTA) and sulfate was determined to 1.45A resolution. The hexameric structure of
SsMTAPII is a dimer-of-trimers with one active site per monomer. The oligomeric
assembly of the trimer and the monomer topology of SsMTAPII are almost identical
with trimeric human 5'-deoxy-5'-methylthioadenosine phosphorylase (hMTAP).
SsMTAPII is the first reported hexameric member in the trimeric class of purine
nucleoside phosphorylase (PNP) from Archaea. Unlike hMTAP, which is highly
specific for MTA, SsMTAPII also accepts adenosine as a substrate. The residues
at the active sites of SsMTAPII and hMTAP are almost identical. The broad
substrate specificity of SsMTAPII may be due to the flexibility of the
C-terminal loop. SsMTAPII is extremely thermoactive and thermostable. The
three-dimensional structure of SsMTAPII suggests that the unique
dimer-of-trimers quaternary structure, a CXC motif at the C terminus, and two
pairs of intrasubunit disulfide bridges may play an important role in its
thermal stability.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Structure of the SsMTAPII subunit. (a) A ribbon
diagram representing the secondary structural elements in the
SsMTAPII subunit (a helices, blue; b strands, green). The
substrate MTA and sulfate are shown in ball-and-stick
representation. The disordered loop composed of residues 255-261
is shown as a dotted line. (b) Topology diagram. The a helices
are shown as blue rectangles, and the b strands are shown as
green arrows. Each secondary structural element is labeled in
its center with its designator and with its beginning and ending
sequence number.
|
 |
Figure 5.
Figure 5. Schematic representation of the SsMTAPII active
site. Residues of SsMTAPII are shown in black boxes. Residues in
red boxes identify structurally equivalent residues in human
MTAP. Residues in blue boxes represent structurally equivalent
residues in human PNP. An asterisk designates residues from the
neighboring subunit. Key hydrogen bonds are indicated by broken
lines with the corresponding donor-acceptor distance labeled.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
357,
252-262)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Cacciapuoti,
I.Peluso,
F.Fuccio,
and
M.Porcelli
(2009).
Purine nucleoside phosphorylases from hyperthermophilic Archaea require a CXC motif for stability and folding.
|
| |
FEBS J,
276,
5799-5805.
|
 |
|
|
|
|
 |
M.Porcelli,
L.Concilio,
I.Peluso,
A.Marabotti,
A.Facchiano,
and
G.Cacciapuoti
(2008).
Pyrimidine-specific ribonucleoside hydrolase from the archaeon Sulfolobus solfataricus--biochemical characterization and homology modeling.
|
| |
FEBS J,
275,
1900-1914.
|
 |
|
|
|
|
 |
P.H.Zwart,
R.W.Grosse-Kunstleve,
A.A.Lebedev,
G.N.Murshudov,
and
P.D.Adams
(2008).
Surprises and pitfalls arising from (pseudo)symmetry.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
99.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Gorassini,
M.F.Mazzeo,
R.A.Siciliano,
V.Carbone,
V.Zappia,
and
M.Porcelli
(2007).
Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus.
|
| |
FEBS J,
274,
2482-2495.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

