 |
PDBsum entry 2a4m

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.2
- tryptophan--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Trp) + L-tryptophan + ATP = L-tryptophyl-tRNA(Trp) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
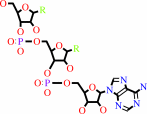 tRNA(Trp)
tRNA(Trp)
|
+
|
L-tryptophan
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
L-tryptophyl-tRNA(Trp)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:31965-31973
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of tryptophanyl-tRNA synthetase II from Deinococcus radiodurans bound to ATP and tryptophan. Insight into subunit cooperativity and domain motions linked to catalysis.
|
|
M.R.Buddha,
B.R.Crane.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
An auxiliary tryptophanyl tRNA synthetase (drTrpRS II) that interacts with
nitric-oxide synthase in the radiation-resistant bacterium Deinococcus
radiodurans charges tRNA with tryptophan and 4-nitrotryptophan, a specific
nitration product of nitric-oxide synthase. Crystal structures of drTrpRS II,
empty of ligands or bound to either Trp or ATP, reveal that drTrpRS II has an
overall structure similar to standard bacterial TrpRSs but undergoes smaller
amplitude motions of the helical tRNA anti-codon binding (TAB) domain on binding
substrates. TAB domain loop conformations that more closely resemble those of
human TrpRS than those of Bacillus stearothermophilus TrpRS (bsTrpRS) indicate
different modes of tRNA recognition by subclasses of bacterial TrpRSs. A compact
state of drTrpRS II binds ATP, from which only minimal TAB domain movement is
necessary to bring nucleotide in contact with Trp. However, the signature KMSKS
loop of class I synthetases does not completely engage the ATP phosphates, and
the adenine ring is not well ordered in the absence of Trp. Thus, progression of
the KMSKS loop to a high energy conformation that stages acyl-adenylation
requires binding of both substrates. In an asymmetric drTrpRS II dimer, the
closed subunit binds ATP, whereas the open subunit binds Trp. A
crystallographically symmetric dimer binds no ligands. Half-site reactivity for
Trp binding is confirmed by thermodynamic measurements and explained by an
asymmetric shift of the dimer interface toward the occupied active site. Upon
Trp binding, Asp68 propagates structural changes between subunits by switching
its hydrogen bonding partner from dimer interface residue Tyr139 to active site
residue Arg30. Since TrpRS IIs are resistant to inhibitors of standard TrpRSs,
and pathogens contain drTrpRS II homologs, the structure of drTrpRS II provides
a framework for the design of potentially useful antibiotics.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. A, ribbon diagram of the drTrpRS II asymmetric
dimer that binds ATP. Each subunit has an RF catalytic domain (B
in cyan and C in dark blue) and a TAB domain (B in violet and C
in red). ATP binds only the B subunit, which has a more closed
conformation than the C subunit. B, ribbon diagram of drTrpRS II
asymmetric dimer colored as in A with L-tryptophan bound in the
more "open" C subunit (Protein Data Bank code 1YI8 [PDB]
). C, superposition of drTrpRS II with bsTrpRS in the open and
closed conformations (Protein Data Bank codes 1D2R [PDB]
and 1I6M, respectively). Only equivalent secondary structure
elements in the respective RF domains were superimposed. D,
superposition of drTrpRS II with human TrpRS (hTrpRS; Protein
Data Bank code 1Q5T [PDB]
).
|
 |
Figure 6.
FIG. 6. A, ATP recognition by bsTrpRS and drTrpRS II. A,
ATP binding site of bsTrpRS in the open conformation (Protein
Data Bank code 1MAW [PDB]
). B, ATP binding site of drTrpRS II with F[o] - F[c] electron
density map (red, 3.5;  , green, 2.5 , green, 2.5  )
calculated with Mg-ATP removed from F[c]. C, active site of
bsTrpRS in closed pretransition state with ATP bound (Protein
Data Bank code 1M83 [PDB]
). In both bsTrpRS and drTrpRS II, magnesium is solely
coordinated by the phosphates of ATP. )
calculated with Mg-ATP removed from F[c]. C, active site of
bsTrpRS in closed pretransition state with ATP bound (Protein
Data Bank code 1M83 [PDB]
). In both bsTrpRS and drTrpRS II, magnesium is solely
coordinated by the phosphates of ATP.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
31965-31973)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.R.Crane,
J.Sudhamsu,
and
B.A.Patel
(2010).
Bacterial nitric oxide synthases.
|
| |
Annu Rev Biochem,
79,
445-470.
|
 |
|
|
|
|
 |
G.W.Han,
X.L.Yang,
D.McMullan,
Y.E.Chong,
S.S.Krishna,
C.L.Rife,
D.Weekes,
S.M.Brittain,
P.Abdubek,
E.Ambing,
T.Astakhova,
H.L.Axelrod,
D.Carlton,
J.Caruthers,
H.J.Chiu,
T.Clayton,
L.Duan,
J.Feuerhelm,
J.C.Grant,
S.K.Grzechnik,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
A.Kumar,
D.Marciano,
M.D.Miller,
A.T.Morse,
E.Nigoghossian,
L.Okach,
J.Paulsen,
R.Reyes,
H.van den Bedem,
A.White,
G.Wolf,
Q.Xu,
K.O.Hodgson,
J.Wooley,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
M.A.Elsliger,
P.Schimmel,
and
I.A.Wilson
(2010).
Structure of a tryptophanyl-tRNA synthetase containing an iron-sulfur cluster.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1326-1334.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Sudhamsu,
and
B.R.Crane
(2009).
Bacterial nitric oxide synthases: what are they good for?
|
| |
Trends Microbiol,
17,
212-218.
|
 |
|
|
|
|
 |
S.Kamijo,
A.Fujii,
K.Onodera,
and
K.Wakabayashi
(2009).
Analyses of conditions for KMSSS loop in tyrosyl-tRNA synthetase by building a mutant library.
|
| |
J Biochem,
146,
241-250.
|
 |
|
|
|
|
 |
W.Tsuchiya,
and
T.Hasegawa
(2009).
Molecular recognition of tryptophan tRNA by tryptophanyl-tRNA synthetase from Aeropyrum pernix K1.
|
| |
J Biochem,
145,
635-641.
|
 |
|
|
|
|
 |
X.L.Yang,
M.Guo,
M.Kapoor,
K.L.Ewalt,
F.J.Otero,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2007).
Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase.
|
| |
Structure,
15,
793-805.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

