 |
PDBsum entry 2a0x

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
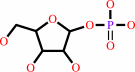 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
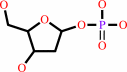 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
46:5038-5049
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
|
|
A.S.Murkin,
M.R.Birck,
A.Rinaldo-Matthis,
W.Shi,
E.A.Taylor,
S.C.Almo,
V.L.Schramm.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystal structures of human purine nucleoside phosphorylase (PNP) with
bound inosine or transition-state analogues show His257 within hydrogen bonding
distance of the 5'-hydroxyl. The mutants His257Phe, His257Gly, and His257Asp
exhibited greatly decreased affinity for Immucillin-H (ImmH), binding this mimic
of an early transition state as much as 370-fold (Km/Ki) less tightly than
native PNP. In contrast, these mutants bound DADMe-ImmH, a mimic of a late
transition state, nearly as well as the native enzyme. These results indicate
that His257 serves an important role in the early stages of transition-state
formation. Whereas mutation of His257 resulted in little variation in the PNP x
DADMe-ImmH x SO4 structures, His257Phe x ImmH x PO4 showed distortion at the
5'-hydroxyl, indicating the importance of H-bonding in positioning this group
during progression to the transition state. Binding isotope effect (BIE) and
kinetic isotope effect (KIE) studies of the remote 5'-(3)H for the arsenolysis
of inosine with native PNP revealed a BIE of 1.5% and an unexpectedly large
intrinsic KIE of 4.6%. This result is interpreted as a moderate electronic
distortion toward the transition state in the Michaelis complex with continued
development of a similar distortion at the transition state. The mutants
His257Phe, His257Gly, and His257Asp altered the 5'-(3)H intrinsic KIE to -3,
-14, and 7%, respectively, while the BIEs contributed 2, 2, and -2%,
respectively. These surprising results establish that forces in the Michaelis
complex, reported by the BIEs, can be reversed or enhanced at the transition
state.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.J.Gainsford,
R.H.Furneaux,
P.C.Tyler,
A.Sauve,
and
V.L.Shramm
(2010).
A synchrotron radiation study of the one-dimensional complex of sodium with (1S)-N-carboxylato-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-D-ribitol, a member of the 'immucillin' family.
|
| |
Acta Crystallogr C,
66,
m65-m68.
|
 |
|
|
|
|
 |
M.C.Ho,
W.Shi,
A.Rinaldo-Matthis,
P.C.Tyler,
G.B.Evans,
K.Clinch,
S.C.Almo,
and
V.L.Schramm
(2010).
Four generations of transition-state analogues for human purine nucleoside phosphorylase.
|
| |
Proc Natl Acad Sci U S A,
107,
4805-4812.
|
 |
|
|
|
|
 |
K.Clinch,
G.B.Evans,
R.F.Fröhlich,
R.H.Furneaux,
P.M.Kelly,
L.Legentil,
A.S.Murkin,
L.Li,
V.L.Schramm,
P.C.Tyler,
and
A.D.Woolhouse
(2009).
Third-generation immucillins: syntheses and bioactivities of acyclic immucillin inhibitors of human purine nucleoside phosphorylase.
|
| |
J Med Chem,
52,
1126-1143.
|
 |
|
|
|
|
 |
M.E.Soliman,
G.D.Ruggiero,
J.J.Pernía,
I.R.Greig,
and
I.H.Williams
(2009).
Computational mutagenesis reveals the role of active-site tyrosine in stabilising a boat conformation for the substrate: QM/MM molecular dynamics studies of wild-type and mutant xylanases.
|
| |
Org Biomol Chem,
7,
460-468.
|
 |
|
|
|
|
 |
M.Ghanem,
A.S.Murkin,
and
V.L.Schramm
(2009).
Ribocation transition state capture and rebound in human purine nucleoside phosphorylase.
|
| |
Chem Biol,
16,
971-979.
|
 |
|
|
|
|
 |
N.R.McIntyre,
E.W.Lowe,
and
D.J.Merkler
(2009).
Imino-oxy acetic acid dealkylation as evidence for an inner-sphere alcohol intermediate in the reaction catalyzed by peptidylglycine alpha-hydroxylating monooxygenase.
|
| |
J Am Chem Soc,
131,
10308-10319.
|
 |
|
|
|
|
 |
S.D.Schwartz,
and
V.L.Schramm
(2009).
Enzymatic transition states and dynamic motion in barrier crossing.
|
| |
Nat Chem Biol,
5,
551-558.
|
 |
|
|
|
|
 |
A.Rinaldo-Matthis,
A.S.Murkin,
U.A.Ramagopal,
K.Clinch,
S.P.Mee,
G.B.Evans,
P.C.Tyler,
R.H.Furneaux,
S.C.Almo,
and
V.L.Schramm
(2008).
L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase.
|
| |
J Am Chem Soc,
130,
842-844.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.S.Murkin,
K.Clinch,
J.M.Mason,
P.C.Tyler,
and
V.L.Schramm
(2008).
Immucillins in custom catalytic-site cavities.
|
| |
Bioorg Med Chem Lett,
18,
5900-5903.
|
 |
|
|
|
|
 |
S.Saen-Oon,
M.Ghanem,
V.L.Schramm,
and
S.D.Schwartz
(2008).
Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase.
|
| |
Biophys J,
94,
4078-4088.
|
 |
|
|
|
|
 |
S.Saen-Oon,
S.Quaytman-Machleder,
V.L.Schramm,
and
S.D.Schwartz
(2008).
Atomic detail of chemical transformation at the transition state of an enzymatic reaction.
|
| |
Proc Natl Acad Sci U S A,
105,
16543-16548.
|
 |
|
|
|
|
 |
S.Saen-Oon,
V.L.Schramm,
and
S.D.Schwartz
(2008).
Transition Path Sampling Study of the Reaction Catalyzed by Purine Nucleoside Phosphorylase.
|
| |
Z Phys Chem (N F),
222,
1359-1374.
|
 |
|
|
|
|
 |
A.Schwarz,
L.Brecker,
and
B.Nidetzky
(2007).
Probing the active site of Corynebacterium callunae starch phosphorylase through the characterization of wild-type and His334-->Gly mutant enzymes.
|
| |
FEBS J,
274,
5105-5115.
|
 |
|
|
|
|
 |
V.L.Schramm
(2007).
Binding isotope effects: boon and bane.
|
| |
Curr Opin Chem Biol,
11,
529-536.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

