 |
PDBsum entry 2zh6

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/RNA
|
PDB id
|
|

|

|
2zh6
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.72
- Cca tRNA nucleotidyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a tRNA precursor + 2 CTP + ATP = a tRNA with a 3' CCA end + 3 diphosphate
|
 |
 |
 |
 |
 |
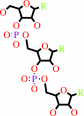 tRNA precursor
tRNA precursor
|
+
|
2
×
CTP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
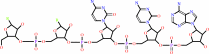 tRNA with a 3' CCA end
tRNA with a 3' CCA end
|
+
|
 3
×
diphosphate
3
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Embo J
27:1944-1952
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme.
|
|
Y.Toh,
T.Numata,
K.Watanabe,
D.Takeshita,
O.Nureki,
K.Tomita.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CCA-adding enzyme builds the 3'-end CCA of tRNA without a nucleic acid template.
The mechanism for the maintenance of fidelity during the CCA-adding reaction
remains elusive. Here, we present almost a dozen complex structures of the class
I CCA-adding enzyme and tRNA mini-helices (mini-D(73)N(74), mini-D(73)N(74)C(75)
and mini-D(73)C(74)N(75); D(73) is a discriminator nucleotide and N is either A,
G, or U). The mini-D(73)N(74) complexes adopt catalytically inactive open forms,
and CTP shifts the enzymes to the active closed forms and allows N(74) to flip
for CMP incorporation. In contrast, unlike the catalytically active closed form
of the mini-D(73)C(74)C(75) complex, the mini-D(73)N(74)C(75) and
mini-D(73)C(74)N(75) complexes adopt inactive open forms. Only the
mini-D(73)C(74)U(75) accepts AMP to a similar extent as mini-D(73)C(74)C(75),
and ATP shifts the enzyme to a closed, active form and allows U(75) to flip for
AMP incorporation. These findings suggest that the 3'-region of RNA is
proofread, after two nucleotide additions, in the closed, active form of the
complex at the AMP incorporation stage. This proofreading is a prerequisite for
the maintenance of fidelity for complete CCA synthesis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Pan,
Y.Xiong,
and
T.A.Steitz
(2010).
How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA.
|
| |
Science,
330,
937-940.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Betat,
C.Rammelt,
and
M.Mörl
(2010).
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.
|
| |
Cell Mol Life Sci,
67,
1447-1463.
|
 |
|
|
|
|
 |
I.U.Heinemann,
D.Söll,
and
L.Randau
(2010).
Transfer RNA processing in archaea: unusual pathways and enzymes.
|
| |
FEBS Lett,
584,
303-309.
|
 |
|
|
|
|
 |
Y.M.Hou
(2010).
CCA addition to tRNA: implications for tRNA quality control.
|
| |
IUBMB Life,
62,
251-260.
|
 |
|
|
|
|
 |
S.Kim,
C.Liu,
K.Halkidis,
H.B.Gamper,
and
Y.M.Hou
(2009).
Distinct kinetic determinants for the stepwise CCA addition to tRNA.
|
| |
RNA,
15,
1827-1836.
|
 |
|
|
|
|
 |
Y.Toh,
D.Takeshita,
T.Numata,
S.Fukai,
O.Nureki,
and
K.Tomita
(2009).
Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme.
|
| |
EMBO J,
28,
3353-3365.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

