 |
PDBsum entry 2zc0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.44
- alanine--glyoxylate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glyoxylate + L-alanine = glycine + pyruvate
|
 |
 |
 |
 |
 |
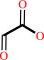 glyoxylate
glyoxylate
|
+
|
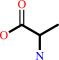 L-alanine
L-alanine
|
=
|
glycine
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PMP)
matches with 88.24% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
64:696-699
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of an archaeal alanine:glyoxylate aminotransferase.
|
|
H.Sakuraba,
K.Yoneda,
K.Takeuchi,
H.Tsuge,
N.Katunuma,
T.Ohshima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of a novel alanine:glyoxylate aminotransferase from the
hyperthermophilic archaeon Thermococcus litoralis was determined at 2.3 A
resolution. The asymmetric unit contains four homologous subunits and the
functional tetramer is generated by noncrystallographic 222 symmetry. Although
the main-chain coordinates of the monomer of the Thermococcus litoralis enzyme
showed a high degree of similarity to those of aspartate aminotransferase from
Thermus thermophilus HB8, the amino-acid residues involved in substrate binding
in the aspartate aminotransferase are only partially conserved in the
Thermococcus litoralis enzyme. This may account for the difference in the
substrate specificities of the two enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2 Stereographic close-up of the substrate-binding site.
The structure of Tc. litoralis AGT (green) is superimposed on
that of Tm. thermophilus AspAT (yellow). The maleate molecule is
shown as a stick model in cyan. The putative interactions are
shown by dotted lines. N and O atoms are shown in blue and red,
respectively.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2008,
64,
696-699)
copyright 2008.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Kameya,
H.Arai,
M.Ishii,
and
Y.Igarashi
(2010).
Purification of three aminotransferases from Hydrogenobacter thermophilus TK-6--novel types of alanine or glycine aminotransferase: enzymes and catalysis.
|
| |
FEBS J,
277,
1876-1885.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

