
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/transferase
|
 |
|
Title:
|
 |
Xenon-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-coa synthase(codh/acs) from moorella thermoacetica
|
|
Structure:
|
 |
Carbon monoxide dehydrogenase/acetyl coa synthase subunit beta. Chain: a, b, c, d. Synonym: carbon monoxide dehydrogenase subunit, codh/acs, codh subunit. Carbon monoxide dehydrogenase/acetyl coa synthase subunit alpha. Chain: m, n, o, p. Synonym: acetyl coa synthase subunit, codh/acs, acs subunit.
|
|
Source:
|
 |
Moorella thermoacetica. Organism_taxid: 1525. Organism_taxid: 1525
|
|
Resolution:
|
 |
|
2.51Å
|
R-factor:
|
0.182
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
T.I.Doukov,L.C.Blasiak,C.L.Drennan
|
|
Key ref:
|
 |
T.I.Doukov
et al.
(2008).
Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry,
47,
3474-3483.
PubMed id:
|
 |
|
Date:
|
 |
|
12-Sep-07
|
Release date:
|
11-Mar-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.1.2.7.4
- anaerobic carbon-monoxide dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
CO + 2 oxidized [2Fe-2S]-[ferredoxin] + H2O = 2 reduced [2Fe-2S]- [ferredoxin] + CO2 + 2 H+
|
 |
 |
 |
 |
 |
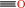 CO
CO
|
+
|
2
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
H2O
|
=
|
2
×
reduced [2Fe-2S]- [ferredoxin]
|
+
|
 CO2
CO2
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation; Ni(2+); Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains M, N, O, P:
E.C.2.3.1.169
- CO-methylating acetyl-CoA synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Co(I)-[corrinoid Fe-S protein] + acetyl-CoA + H+ = methyl-Co(III)- [corrinoid Fe-S protein] + CO + CoA
|
 |
 |
 |
 |
 |
Co(I)-[corrinoid Fe-S protein]
|
+
|
 2
×
acetyl-CoA
2
×
acetyl-CoA
|
+
|
H(+)
|
=
|
2
×
methyl-Co(III)- [corrinoid Fe-S protein]
|
+
|
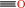 CO
CO
|
+
|
 2
×
CoA
2
×
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu(2+); Iron-sulfur; Ni(2+)
|
 |
 |
 |
 |
 |
Cu(2+)
|
 Iron-sulfur
Iron-sulfur
|
Ni(2+)
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:3474-3483
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
|
|
T.I.Doukov,
L.C.Blasiak,
J.Seravalli,
S.W.Ragsdale,
C.L.Drennan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A fascinating feature of some bifunctional enzymes is the presence of an
internal channel or tunnel to connect the multiple active sites. A channel can
allow for a reaction intermediate generated at one active site to be used as a
substrate at a second active site, without the need for the intermediate to
leave the safety of the protein matrix. One such bifunctional enzyme is carbon
monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica
(mtCODH/ACS). A key player in the global carbon cycle, CODH/ACS uses a Ni-Fe-S
center called the C-cluster to reduce carbon dioxide to carbon monoxide and uses
a second Ni-Fe-S center, called the A-cluster, to assemble acetyl-CoA from a
methyl group, coenzyme A, and C-cluster-generated CO. mtCODH/ACS has been
proposed to contain one of the longest enzyme channels (138 A long) to allow for
intermolecular CO transport. Here, we report a 2.5 A resolution structure of
xenon-pressurized mtCODH/ACS and examine the nature of gaseous cavities within
this enzyme. We find that the cavity calculation program CAVENV accurately
predicts the channels connecting the C- and A-clusters, with 17 of 19 xenon
binding sites within the predicted regions. Using this X-ray data, we analyze
the amino acid composition surrounding the 19 Xe sites and consider how the
protein fold is utilized to carve out such an impressive interior passageway.
Finally, structural comparisons of Xe-pressurized mtCODH/ACS with related enzyme
structures allow us to study channel design principles, as well as consider the
conformational flexibility of an enzyme that contains a cavity through its
center.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Kung,
and
C.L.Drennan
(2011).
A role for nickel-iron cofactors in biological carbon monoxide and carbon dioxide utilization.
|
| |
Curr Opin Chem Biol,
15,
276-283.
|
 |
|
|
|
|
 |
A.S.Olia,
S.Casjens,
and
G.Cingolani
(2009).
Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas.
|
| |
Protein Sci,
18,
537-548.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Volbeda,
C.Darnault,
X.Tan,
P.A.Lindahl,
and
J.C.Fontecilla-Camps
(2009).
Novel domain arrangement in the crystal structure of a truncated acetyl-CoA synthase from Moorella thermoacetica.
|
| |
Biochemistry,
48,
7916-7926.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.M.Barney,
M.G.Yurth,
P.C.Dos Santos,
D.R.Dean,
and
L.C.Seefeldt
(2009).
A substrate channel in the nitrogenase MoFe protein.
|
| |
J Biol Inorg Chem,
14,
1015-1022.
|
 |
|
|
|
|
 |
J.C.Fontecilla-Camps,
P.Amara,
C.Cavazza,
Y.Nicolet,
and
A.Volbeda
(2009).
Structure-function relationships of anaerobic gas-processing metalloenzymes.
|
| |
Nature,
460,
814-822.
|
 |
|
|
|
|
 |
M.D.Fayer
(2009).
Dynamics of liquids, molecules, and proteins measured with ultrafast 2D IR vibrational echo chemical exchange spectroscopy.
|
| |
Annu Rev Phys Chem,
60,
21-38.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2009).
Nickel-based Enzyme Systems.
|
| |
J Biol Chem,
284,
18571-18575.
|
 |
|
|
|
|
 |
Y.Kung,
T.I.Doukov,
J.Seravalli,
S.W.Ragsdale,
and
C.L.Drennan
(2009).
Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
|
| |
Biochemistry,
48,
7432-7440.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Ishikawa,
K.Kwak,
J.K.Chung,
S.Kim,
and
M.D.Fayer
(2008).
Direct observation of fast protein conformational switching.
|
| |
Proc Natl Acad Sci U S A,
105,
8619-8624.
|
 |
|
|
|
|
 |
J.W.Murray,
K.Maghlaoui,
J.Kargul,
M.Sugiura,
and
J.Barber
(2008).
Analysis of xenon binding to photosystem II by X-ray crystallography.
|
| |
Photosynth Res,
98,
523-527.
|
 |
|
|
|
|
 |
S.W.Ragsdale,
and
E.Pierce
(2008).
Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation.
|
| |
Biochim Biophys Acta,
1784,
1873-1898.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2008).
Catalysis of methyl group transfers involving tetrahydrofolate and B(12).
|
| |
Vitam Horm,
79,
293-324.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

