 |
PDBsum entry 2z7b

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.51
- 3-hydroxy-2-methylpyridine-4,5-dicarboxylate 4-decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-hydroxy-6-methylpyridine-3,4-dicarboxylate + H+ = 3-hydroxy-2- methylpyridine-5-carboxylate + CO2
|
 |
 |
 |
 |
 |
5-hydroxy-6-methylpyridine-3,4-dicarboxylate
|
+
|
H(+)
|
=
|
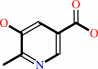 3-hydroxy-2- methylpyridine-5-carboxylate
3-hydroxy-2- methylpyridine-5-carboxylate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
46:13606-13615
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Gene identification and structural characterization of the pyridoxal 5'-phosphate degradative protein 3-hydroxy-2-methylpyridine-4,5-dicarboxylate decarboxylase from mesorhizobium loti MAFF303099.
|
|
T.Mukherjee,
K.M.McCulloch,
S.E.Ealick,
T.P.Begley.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The function of the mlr6791 gene from Mesorhizobium loti MAFF303099 has been
identified. This gene encodes 3-hydroxy-2-methylpyridine-4,5-dicarboxylate
decarboxylase (HMPDdc), an enzyme involved in the catabolism of pyridoxal
5'-phosphate (Vitamin B6). This enzyme was overexpressed in Escherichia coli and
characterized. HMPDdc is a 26 kDa protein that catalyzes the decarboxylation of
3-hydroxy-2-methylpyridine-4,5-dicarboxylate to
3-hydroxy-2-methylpyridine-5-carboxylate. The KM and kcat were found to be 366
microM and 0.6 s-1, respectively. The structure of this enzyme was determined at
1.9 A resolution using SAD phasing and belongs to the class II aldolase/adducin
superfamily. While the decarboxylation of hydroxy-substituted benzene rings is a
common motif in biosynthesis, the mechanism of this reaction is still poorly
characterized. The structural studies described here suggest that catalysis of
such decarboxylations proceeds by an aldolase-like mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.M.McCulloch,
T.Mukherjee,
T.P.Begley,
and
S.E.Ealick
(2010).
Structure determination and characterization of the vitamin B6 degradative enzyme (E)-2-(acetamidomethylene)succinate hydrolase.
|
| |
Biochemistry,
49,
1226-1235.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.M.McCulloch,
T.Mukherjee,
T.P.Begley,
and
S.E.Ealick
(2009).
Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
|
| |
Biochemistry,
48,
4139-4149.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Yokochi,
Y.Yoshikane,
S.Matsumoto,
M.Fujisawa,
K.Ohnishi,
and
T.Yagi
(2009).
Gene identification and characterization of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD+-dependent dismutase.
|
| |
J Biochem,
145,
493-503.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

