 |
PDBsum entry 2z3l

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.2.6
- lysine/arginine leucyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N-terminal L-lysyl-[protein] + L-leucyl-tRNA(Leu) = N-terminal L-leucyl-L-lysyl-[protein] + tRNA(Leu) + H(+)
|
|
2.
|
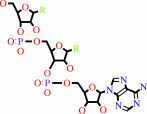
N-terminal L-arginyl-[protein] + L-leucyl-tRNA(Leu) = N-terminal L-leucyl-L-arginyl-[protein] + tRNA(Leu) + H(+)
|
|
 |
 |
 |
 |
 |
N-terminal L-lysyl-[protein]
|
+
|
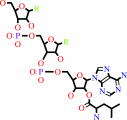 L-leucyl-tRNA(Leu)
L-leucyl-tRNA(Leu)
|
=
|
N-terminal L-leucyl-L-lysyl-[protein]
|
+
|
 tRNA(Leu)
tRNA(Leu)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
N-terminal L-arginyl-[protein]
|
+
|
 L-leucyl-tRNA(Leu)
L-leucyl-tRNA(Leu)
|
=
|
N-terminal L-leucyl-L-arginyl-[protein]
|
+
|
 tRNA(Leu)
tRNA(Leu)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Monovalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
449:867-871
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase.
|
|
K.Watanabe,
Y.Toh,
K.Suto,
Y.Shimizu,
N.Oka,
T.Wada,
K.Tomita.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Eubacterial leucyl/phenylalanyl-tRNA protein transferase (LF-transferase)
catalyses peptide-bond formation by using Leu-tRNA(Leu) (or Phe-tRNA(Phe)) and
an amino-terminal Arg (or Lys) of a protein, as donor and acceptor substrates,
respectively. However, the catalytic mechanism of peptide-bond formation by
LF-transferase remained obscure. Here we determine the structures of complexes
of LF-transferase and phenylalanyl adenosine, with and without a short peptide
bearing an N-terminal Arg. Combining the two separate structures into one
structure as well as mutation studies reveal the mechanism for peptide-bond
formation by LF-transferase. The electron relay from Asp 186 to Gln 188 helps
Gln 188 to attract a proton from the alpha-amino group of the N-terminal Arg of
the acceptor peptide. This generates the attacking nucleophile for the carbonyl
carbon of the aminoacyl bond of the aminoacyl-tRNA, thus facilitating
peptide-bond formation. The protein-based mechanism for peptide-bond formation
by LF-transferase is similar to the reverse reaction of the acylation step
observed in the peptide hydrolysis reaction by serine proteases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3: Superposition of two binary complex structures. a,
Structural difference between the complex with rA-Phe (blue) and
that with the product peptide (green). b, Detailed structural
difference of the Gln 188 side chains between the two
structures. c, Superposition of the two complexes. The complex
with rA-Phe and that with the product peptide are coloured blue
and dark green, respectively. The benzyl group of rA-Phe and the
Arg in the product peptide are coloured cyan and green,
respectively, and are shown in stick models.
|
 |
Figure 4.
Figure 4: A model of the catalytic mechanism for peptide-bond
formation by LF-transferase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2007,
449,
867-871)
copyright 2007.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Bonnefond,
T.Arai,
Y.Sakaguchi,
T.Suzuki,
R.Ishitani,
and
O.Nureki
(2011).
Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
|
| |
Proc Natl Acad Sci U S A,
108,
3912-3917.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.M.Sriram,
B.Y.Kim,
and
Y.T.Kwon
(2011).
The N-end rule pathway: emerging functions and molecular principles of substrate recognition.
|
| |
Nat Rev Mol Cell Biol,
12,
735-747.
|
 |
|
|
|
|
 |
G.Lahoud,
and
Y.M.Hou
(2010).
Biosynthesis: A new (old) way of hijacking tRNA.
|
| |
Nat Chem Biol,
6,
795-796.
|
 |
|
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
T.Yanagisawa,
T.Sumida,
R.Ishii,
C.Takemoto,
and
S.Yokoyama
(2010).
A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
|
| |
Nat Struct Mol Biol,
17,
1136-1143.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.von Döhren
(2009).
Charged tRNAs charge into secondary metabolism.
|
| |
Nat Chem Biol,
5,
374-375.
|
 |
|
|
|
|
 |
J.Kirstein,
N.Molière,
D.A.Dougan,
and
K.Turgay
(2009).
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.
|
| |
Nat Rev Microbiol,
7,
589-599.
|
 |
|
|
|
|
 |
J.Ling,
B.R.So,
S.S.Yadavalli,
H.Roy,
S.Shoji,
K.Fredrick,
K.Musier-Forsyth,
and
M.Ibba
(2009).
Resampling and editing of mischarged tRNA prior to translation elongation.
|
| |
Mol Cell,
33,
654-660.
|
 |
|
|
|
|
 |
K.Ebisu,
H.Tateno,
H.Kuroiwa,
K.Kawakami,
M.Ikeuchi,
J.Hirabayashi,
M.Sisido,
and
M.Taki
(2009).
N-terminal specific point-immobilization of active proteins by the one-pot NEXT-A method.
|
| |
Chembiochem,
10,
2460-2464.
|
 |
|
|
|
|
 |
M.Fonvielle,
M.Chemama,
R.Villet,
M.Lecerf,
A.Bouhss,
J.M.Valéry,
M.Ethève-Quelquejeu,
and
M.Arthur
(2009).
Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis.
|
| |
Nucleic Acids Res,
37,
1589-1601.
|
 |
|
|
|
|
 |
M.Gondry,
L.Sauguet,
P.Belin,
R.Thai,
R.Amouroux,
C.Tellier,
K.Tuphile,
M.Jacquet,
S.Braud,
M.Courçon,
C.Masson,
S.Dubois,
S.Lautru,
A.Lecoq,
S.Hashimoto,
R.Genet,
and
J.L.Pernodet
(2009).
Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes.
|
| |
Nat Chem Biol,
5,
414-420.
|
 |
|
|
|
|
 |
R.L.Ninnis,
S.K.Spall,
G.H.Talbo,
K.N.Truscott,
and
D.A.Dougan
(2009).
Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli.
|
| |
EMBO J,
28,
1732-1744.
|
 |
|
|
|
|
 |
V.J.Schuenemann,
S.M.Kralik,
R.Albrecht,
S.K.Spall,
K.N.Truscott,
D.A.Dougan,
and
K.Zeth
(2009).
Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS.
|
| |
EMBO Rep,
10,
508-514.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Varshavsky
(2008).
The N-end rule at atomic resolution.
|
| |
Nat Struct Mol Biol,
15,
1238-1240.
|
 |
|
|
|
|
 |
H.Roy,
and
M.Ibba
(2008).
RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors.
|
| |
Proc Natl Acad Sci U S A,
105,
4667-4672.
|
 |
|
|
|
|
 |
M.Taki,
H.Kuroiwa,
and
M.Sisido
(2008).
Chemoenzymatic transfer of fluorescent non-natural amino acids to the N terminus of a protein/peptide.
|
| |
Chembiochem,
9,
719-722.
|
 |
|
|
|
|
 |
U.L.RajBhandary,
and
D.Söll
(2008).
Aminoacyl-tRNAs, the bacterial cell envelope, and antibiotics.
|
| |
Proc Natl Acad Sci U S A,
105,
5285-5286.
|
 |
|
|
|
|
 |
Z.Xia,
A.Webster,
F.Du,
K.Piatkov,
M.Ghislain,
and
A.Varshavsky
(2008).
Substrate-binding Sites of UBR1, the Ubiquitin Ligase of the N-end Rule Pathway.
|
| |
J Biol Chem,
283,
24011-24028.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

