 |
PDBsum entry 2yyt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of uncharacterized conserved protein from geobacillus kaustophilus
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a, b, c, d. Synonym: omp decarboxylase, ompdcase, ompdecase. Engineered: yes
|
|
Source:
|
 |
Geobacillus kaustophilus. Organism_taxid: 235909. Strain: hta426. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.229
|
R-free:
|
0.289
|
|
|
Authors:
|
 |
M.Kanagawa,S.Baba,Y.Nakamura,Y.Bessho,S.Kuramitsu,S.Yokoyama,G.Kawai, G.Sampei,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
M.Kanagawa
et al.
Crystal structure of uncharacterized conserved protein from geobacillus kaustophilus.
To be published,
.
|
 |
|
Date:
|
 |
|
02-May-07
|
Release date:
|
06-Nov-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5L0U0
(PYRF_GEOKA) -
Orotidine 5'-phosphate decarboxylase from Geobacillus kaustophilus (strain HTA426)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
244 a.a.
215 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
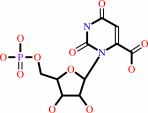 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

