 |
PDBsum entry 2v7n

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Immune system
|
PDB id
|
|

|

|
2v7n
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Immune system
|
 |
|
Title:
|
 |
Unusual twinning in crystals of the cits binding antibody fab fragment f3p4
|
|
Structure:
|
 |
Immunoglobulin light chain. Chain: a, c, e, g. Fragment: fab fragment, residues 1-255. Synonym: vk3. Engineered: yes. Other_details: synthetic gene, derived from phage display of the human combinatorial antibody library (hucal). Immunoglobulin heavy chain. Chain: b, d, f, h.
|
|
Source:
|
 |
Synthetic construct. Organism_taxid: 32630. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.92Å
|
R-factor:
|
0.155
|
R-free:
|
0.202
|
|
|
Authors:
|
 |
D.Frey,T.Huber,A.Plueckthun,M.G.Gruetter
|
Key ref:

|
 |
D.Frey
et al.
(2008).
Structure of the recombinant antibody Fab fragment f3p4.
Acta Crystallogr D Biol Crystallogr,
64,
636-643.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
31-Jul-07
|
Release date:
|
17-Jun-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
64:636-643
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the recombinant antibody Fab fragment f3p4.
|
|
D.Frey,
T.Huber,
A.Plückthun,
M.G.Grütter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the antibody Fab fragment f3p4, which was selected from a
subset of the synthetic HuCAL antibody library to bind the sodium citrate
symporter CitS, is described at 1.92 A resolution. Comparison with computational
models revealed deviations in a few framework positions and in the binding
loops. The crystals belong to space group P2(1)2(1)2 and contain four molecules
in the asymmetric unit, with unit-cell parameters a=102.77, b=185.92, c=102.97
A. These particular unit-cell parameters allowed pseudo-merohedral twinning;
interestingly, the twinning law relates a twofold screw axis to a twofold axis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5 Structure of CDR-L3. A superposition of the main-chain
atoms (for clarity, carbonyl O atoms are omitted from the
figure) of CDR-L3 of the Fab fragment f3p4 (grey) with typical
members of the V[  ]and
V[ ]and
V[ 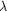 ]families
is shown. In V[ ]families
is shown. In V[ 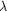 ]members
CDR-L3 adopts a hairpin-like structure (magenta; PDB code 8fab
). In V[ ]members
CDR-L3 adopts a hairpin-like structure (magenta; PDB code 8fab
). In V[  ]members,
which predominantly have a proline at position L136 (Kabat L95),
the CDR-L3 has an ]members,
which predominantly have a proline at position L136 (Kabat L95),
the CDR-L3 has an  -loop
conformation (yellow; PDB code 1ay1 ). The conformation of the
CDR-L3 of Fab fragment f3p4 is an intermediate conformation, but
is also present in naturally occurring antibodies with a proline
residue at L136 (Kabat L95; green; PDB code 1c1e ). The compared
molecules have an equal number of residues in CDR-L3 and all C^ -loop
conformation (yellow; PDB code 1ay1 ). The conformation of the
CDR-L3 of Fab fragment f3p4 is an intermediate conformation, but
is also present in naturally occurring antibodies with a proline
residue at L136 (Kabat L95; green; PDB code 1c1e ). The compared
molecules have an equal number of residues in CDR-L3 and all C^
 atoms
of L106-L140 (Kabat L88-99) were used for the superposition. atoms
of L106-L140 (Kabat L88-99) were used for the superposition.
|
 |
Figure 7.
Figure 7 Surface representation of the binding region. C^  traces
of CDR-H1, CDR-H2, CDR-H3, CDR-L1, CDR-L2 and CDR-L3 are shown
and coloured red, green, blue, cyan, magenta and yellow,
respectively. Residues Arg H108 (Kabat H94) and Asp H137 (Kabat
H101) forming the conserved hydrogen bonds as well as the
solvent-exposed residue Tyr L40 (Kabat L32) are shown in stick
representation. traces
of CDR-H1, CDR-H2, CDR-H3, CDR-L1, CDR-L2 and CDR-L3 are shown
and coloured red, green, blue, cyan, magenta and yellow,
respectively. Residues Arg H108 (Kabat H94) and Asp H137 (Kabat
H101) forming the conserved hydrogen bonds as well as the
solvent-exposed residue Tyr L40 (Kabat L32) are shown in stick
representation.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2008,
64,
636-643)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.X.Zhao,
L.Yang,
Z.N.Gu,
H.Q.Chen,
F.W.Tian,
Y.Q.Chen,
H.Zhang,
and
W.Chen
(2010).
Stabilization of the single-chain fragment variable by an interdomain disulfide bond and its effect on antibody affinity.
|
| |
Int J Mol Sci,
12,
1.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

