 |
PDBsum entry 2v6b

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2v6b
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.27
- L-lactate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-lactate + NAD+ = pyruvate + NADH + H+
|
 |
 |
 |
 |
 |
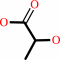 (S)-lactate
(S)-lactate
|
+
|
 NAD(+)
NAD(+)
|
=
|
 pyruvate
pyruvate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
374:547-562
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments.
|
|
N.Coquelle,
E.Fioravanti,
M.Weik,
F.Vellieux,
D.Madern.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Lactate dehydrogenase (LDH) catalyzes the conversion of pyruvate to lactate with
concomitant oxidation of NADH during the last step in anaerobic glycolysis. In
the present study, we present a comparative biochemical and structural analysis
of various LDHs adapted to function over a large temperature range. The enzymes
were from Champsocephalus gunnari (an Antarctic fish), Deinococcus radiodurans
(a mesophilic bacterium) and Thermus thermophilus (a hyperthermophilic
bacterium). The thermodynamic activation parameters of these LDHs indicated that
temperature adaptation from hot to cold conditions was due to a decrease in the
activation enthalpy and an increase in activation entropy. The crystal
structures of these LDHs have been solved. Pairwise comparisons at the
structural level, between hyperthermophilic versus mesophilic LDHs and
mesophilic versus psychrophilic LDHs, have revealed that temperature adaptation
is due to a few amino acid substitutions that are localized in critical regions
of the enzyme. These substitutions, each having accumulating effects, play a
role in either the conformational stability or the local flexibility or in both.
Going from hot- to cold-adapted LDHs, the various substitutions have decreased
the number of ion pairs, reduced the size of ionic networks, created unfavorable
interactions involving charged residues and induced strong local disorder. The
analysis of the LDHs adapted to extreme temperatures shed light on how
evolutionary processes shift the subtle balance between overall stability and
flexibility of an enzyme.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Ribbon diagram of the apo form of tetrameric TtLDH.
The four monomers are labeled A–D.
|
 |
Figure 3.
Fig. 3. A 30 Å slice through a ribbon diagram of a
monomer of TtLDH complexed with oxamate and NADH. The color
scheme used is related to C^α deviation between the apo and
ternary complex. The rainbow scale bar illustrates values of
deviation in angstroms. Inset: RMSD between C^α atoms in the
superimposed subunits of TtLDH (apo versus ternary complex). The
mobile regions (MR) that display the largest deviations are
indicated.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
374,
547-562)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Tiberti,
and
E.Papaleo
(2011).
Dynamic properties of extremophilic subtilisin-like serine-proteases.
|
| |
J Struct Biol,
174,
69-83.
|
 |
|
|
|
|
 |
C.Michaux,
J.Massant,
F.Kerff,
J.M.Frère,
J.D.Docquier,
I.Vandenberghe,
B.Samyn,
A.Pierrard,
G.Feller,
P.Charlier,
J.Van Beeumen,
and
J.Wouters
(2008).
Crystal structure of a cold-adapted class C beta-lactamase.
|
| |
FEBS J,
275,
1687-1697.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.F.Rodrigues,
and
J.M.Tiedje
(2008).
Coping with our cold planet.
|
| |
Appl Environ Microbiol,
74,
1677-1686.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

