 |
PDBsum entry 2uya

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.2
- oxalate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxalate + H+ = formate + CO2
|
 |
 |
 |
 |
 |
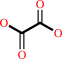 oxalate
oxalate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
 formate
formate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
407:397-406
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The identity of the active site of oxalate decarboxylase and the importance of the stability of active-site lid conformations.
|
|
V.J.Just,
M.R.Burrell,
L.Bowater,
I.McRobbie,
C.E.Stevenson,
D.M.Lawson,
S.Bornemann.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Oxalate decarboxylase (EC 4.1.1.2) catalyses the conversion of oxalate into
carbon dioxide and formate. It requires manganese and, uniquely, dioxygen for
catalysis. It forms a homohexamer and each subunit contains two similar, but
distinct, manganese sites termed sites 1 and 2. There is kinetic evidence that
only site 1 is catalytically active and that site 2 is purely structural.
However, the kinetics of enzymes with mutations in site 2 are often ambiguous
and all mutant kinetics have been interpreted without structural information.
Nine new site-directed mutants have been generated and four mutant crystal
structures have now been solved. Most mutants targeted (i) the flexibility
(T165P), (ii) favoured conformation (S161A, S164A, D297A or H299A) or (iii)
presence (Delta162-163 or Delta162-164) of a lid associated with site 1. The
kinetics of these mutants were consistent with only site 1 being catalytically
active. This was particularly striking with D297A and H299A because they
disrupted hydrogen bonds between the lid and a neighbouring subunit only when in
the open conformation and were distant from site 2. These observations also
provided the first evidence that the flexibility and stability of lid
conformations are important in catalysis. The deletion of the lid to mimic the
plant oxalate oxidase led to a loss of decarboxylase activity, but only a slight
elevation in the oxalate oxidase side reaction, implying other changes are
required to afford a reaction specificity switch. The four mutant crystal
structures (R92A, E162A, Delta162-163 and S161A) strongly support the hypothesis
that site 2 is purely structural.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Imaram,
B.T.Saylor,
C.P.Centonze,
N.G.Richards,
and
A.Angerhofer
(2011).
EPR spin trapping of an oxalate-derived free radical in the oxalate decarboxylase reaction.
|
| |
Free Radic Biol Med,
50,
1009-1015.
|
 |
|
|
|
|
 |
M.R.Mäkelä,
K.Hildén,
and
T.K.Lundell
(2010).
Oxalate decarboxylase: biotechnological update and prevalence of the enzyme in filamentous fungi.
|
| |
Appl Microbiol Biotechnol,
87,
801-814.
|
 |
|
|
|
|
 |
E.W.Moomaw,
A.Angerhofer,
P.Moussatche,
A.Ozarowski,
I.García-Rubio,
and
N.G.Richards
(2009).
Metal dependence of oxalate decarboxylase activity.
|
| |
Biochemistry,
48,
6116-6125.
|
 |
|
|
|
|
 |
L.C.Tabares,
J.Gätjens,
C.Hureau,
M.R.Burrell,
L.Bowater,
V.L.Pecoraro,
S.Bornemann,
and
S.Un
(2009).
pH-dependent structures of the manganese binding sites in oxalate decarboxylase as revealed by high-field electron paramagnetic resonance.
|
| |
J Phys Chem B,
113,
9016-9025.
|
 |
|
|
|
|
 |
S.R.MacLellan,
T.Wecke,
and
J.D.Helmann
(2008).
A previously unidentified sigma factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis.
|
| |
Mol Microbiol,
69,
954-967.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

