 |
PDBsum entry 2tpt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.4
- thymidine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + phosphate = 2-deoxy-alpha-D-ribose 1-phosphate + thymine
|
 |
 |
 |
 |
 |
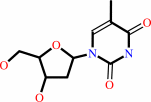 thymidine
thymidine
|
+
|
 phosphate
phosphate
|
=
|
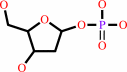 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
+
|
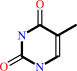 thymine
thymine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
281:285-299
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
|
|
M.J.Pugmire,
W.J.Cook,
A.Jasanoff,
M.R.Walter,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Two new crystal forms of Escherichia coli thymidine phosphorylase (EC 2.4.2.4)
have been found; a monoclinic form (space group P21) and an orthorhombic form
(space group I222). These structures have been solved and compared to the
previously determined tetragonal form (space group P43212). This comparison
provides evidence of domain movement of the alpha (residues 1 to 65, 163 to 193)
and alpha/beta (residues 80 to 154, 197 to 440) domains, which is thought to be
critical for enzymatic activity by closing the active site cleft. Three hinge
regions apparently allow the alpha and alpha/beta-domains to move relative to
each other. The monoclinic model is the most open of the three models while the
tetragonal model is the most closed. Phosphate binding induces formation of a
hydrogen bond between His119 and Gly208, which helps to order the 115 to 120
loop that is disordered prior to phosphate binding. The formation of this
hydrogen bond also appears to play a key role in the domain movement. The
alpha-domain moves as a rigid body, while the alpha/beta-domain has some
non-rigid body movement that is associated with the formation of the
His119-Gly208 hydrogen bond. The 8 A distance between the two substrates
reported for the tetragonal form indicates that it is probably not in an active
conformation. However, the structural data for these two new crystal forms
suggest that closing the interdomain cleft around the substrates may generate a
functional active site. Molecular modeling and dynamics simulation techniques
have been used to generate a hypothetical closed conformation of the enzyme.
Analysis of this model suggests several residues of possible catalytic
importance. The model explains observed kinetic results and satisfies
requirements for efficient enzyme catalysis, most notably through the exclusion
of water from the enzyme's active site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. (a) Ribbon drawing of a monomer of the E. coli
thymidine phosphorylase tetragonal crystal structure produced
with the program INSIGHT II, v. 95.0 (Biosym Technologies Inc).
a-Helices are labeled H1 to H17 and are represented as red
cylinders. The two b-sheets are colored yellow and are labeled A
and B. Phosphate and thymine are shown, indicating the location
of the phosphate and thymidine binding sites. (b) Stereoview of
a C^a trace of the TPT dimer, where the dimer axis is
perpendicular to the plane of the page. Labels of every 20th
residue position are included, where ` indicates the
corresponding residue in the second subunit. This Figure was
prodcued using MOLSCRIPT [Kraulis 1991].
|
 |
Figure 2.
Figure 2. Stereoview of the phosphate binding site in the
TPT model. Sulfate ion is likely bound in the phosphate binding
site due to the high concentration of ammonium sulfate in the
crystallization conditions. Possible hydrogen bonds are
indicated by dotted lines. This Figure was prepared using the
program MOLSCRIPT [Kraulis 1991].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1998,
281,
285-299)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Bronckaers,
F.Gago,
J.Balzarini,
and
S.Liekens
(2009).
The dual role of thymidine phosphorylase in cancer development and chemotherapy.
|
| |
Med Res Rev,
29,
903-953.
|
 |
|
|
|
|
 |
K.Shimizu,
and
N.Kunishima
(2007).
Purification, crystallization and preliminary X-ray diffraction study on pyrimidine nucleoside phosphorylase TTHA1771 from Thermus thermophilus HB8.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
308-310.
|
 |
|
|
|
|
 |
N.G.Panova,
C.S.Alexeev,
A.S.Kuzmichov,
E.V.Shcheveleva,
S.A.Gavryushov,
K.M.Polyakov,
A.M.Kritzyn,
S.N.Mikhailov,
R.S.Esipov,
and
A.I.Miroshnikov
(2007).
Substrate specificity of Escherichia coli thymidine phosphorylase.
|
| |
Biochemistry (Mosc),
72,
21-28.
|
 |
|
|
|
|
 |
W.Bu,
E.C.Settembre,
M.H.el Kouni,
and
S.E.Ealick
(2005).
Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
863-872.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.A.Norman,
S.T.Barry,
M.Bate,
J.Breed,
J.G.Colls,
R.J.Ernill,
R.W.Luke,
C.A.Minshull,
M.S.McAlister,
E.J.McCall,
H.H.McMiken,
D.S.Paterson,
D.Timms,
J.A.Tucker,
and
R.A.Pauptit
(2004).
Crystal structure of human thymidine phosphorylase in complex with a small molecule inhibitor.
|
| |
Structure,
12,
75-84.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Mayans,
A.Ivens,
L.J.Nissen,
K.Kirschner,
and
M.Wilmanns
(2002).
Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry.
|
| |
EMBO J,
21,
3245-3254.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.C.Appleby,
I.I.Mathews,
M.Porcelli,
G.Cacciapuoti,
and
S.E.Ealick
(2001).
Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
|
| |
J Biol Chem,
276,
39232-39242.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Ishijima,
T.Nakai,
S.Kawaguchi,
K.Hirotsu,
and
S.Kuramitsu
(2000).
Free energy requirement for domain movement of an enzyme.
|
| |
J Biol Chem,
275,
18939-18945.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Nishino,
A.Spinazzola,
and
M.Hirano
(1999).
Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder.
|
| |
Science,
283,
689-692.
|
 |
|
|
|
|
 |
S.W.Rick,
Y.G.Abashkin,
R.L.Hilderbrandt,
and
S.K.Burt
(1999).
Computational studies of the domain movement and the catalytic mechanism of thymidine phosphorylase.
|
| |
Proteins,
37,
242-252.
|
 |
|
|
|
|
 |
M.J.Pugmire,
and
S.E.Ealick
(1998).
The crystal structure of pyrimidine nucleoside phosphorylase in a closed conformation.
|
| |
Structure,
6,
1467-1479.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

