 |
PDBsum entry 2qlt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of an isoform of dl-glycerol-3-phosphatase, rhr2p, from saccharomyces cerevisiae
|
|
Structure:
|
 |
(Dl)-glycerol-3-phosphatase 1. Chain: a. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Gene: gpp1, rhr2, yil053w. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.169
|
R-free:
|
0.195
|
|
|
Authors:
|
 |
K.Tan,E.Evdokimova,M.Kudritska,A.Savchenko,A.Edwards,A.Joachimiak, Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
K.Tan
et al.
The crystal structure of an isoform of dl-Glycerol-3-Phosphatase, Rhr2p from saccharomyces cerevisiae..
To be published,
.
|
 |
|
Date:
|
 |
|
13-Jul-07
|
Release date:
|
07-Aug-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P41277
(GPP1_YEAST) -
Glycerol-1-phosphate phosphohydrolase 1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
250 a.a.
251 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.21
- glycerol-1-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
sn-glycerol 1-phosphate + H2O = glycerol + phosphate
|
|
2.
|
sn-glycerol 3-phosphate + H2O = glycerol + phosphate
|
|
 |
 |
 |
 |
 |
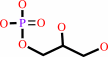 sn-glycerol 1-phosphate
sn-glycerol 1-phosphate
|
+
|
H2O
|
=
|
glycerol
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
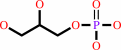 sn-glycerol 3-phosphate
sn-glycerol 3-phosphate
|
+
|
H2O
|
=
|
glycerol
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

