 |
PDBsum entry 2o7u

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal transport
|
PDB id
|
|

|

|
2o7u
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Metal transport
|
 |
|
Title:
|
 |
Crystal structure of k206e/k296e mutant of the n-terminal half molecule of human transferrin
|
|
Structure:
|
 |
Serotransferrin. Chain: b, a, c, d, e, f, g, h, i. Fragment: n-lobe. Synonym: transferrin, siderophilin, beta-1-metal-binding globulin. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: mesocricetus auratus. Expression_system_taxid: 10036. Expression_system_cell_line: bhk. Expression_system_organ: kidney.
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.231
|
R-free:
|
0.259
|
|
|
Authors:
|
 |
H.M.Baker,D.Nurizzo,A.B.Mason,E.N.Baker
|
Key ref:

|
 |
H.M.Baker
et al.
(2007).
Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr D Biol Crystallogr,
63,
408-414.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Dec-06
|
Release date:
|
23-Jan-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P02787
(TRFE_HUMAN) -
Serotransferrin from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
698 a.a.
329 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
63:408-414
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
|
|
H.M.Baker,
D.Nurizzo,
A.B.Mason,
E.N.Baker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Iron uptake by humans depends on the ability of the serum protein transferrin
(Tf) to bind iron as Fe(3+) with high affinity but reversibly. Iron release into
cells occurs through receptor-mediated endocytosis, aided by the lower endosomal
pH of about 5.5. The protonation of a hydrogen-bonded pair of lysines, Lys206
and Lys296, adjacent to the N-lobe iron site of Tf has been proposed to create a
repulsive interaction that stimulates domain opening and iron release. The
crystal structures of two mutants, K206E (in which Lys206 is mutated to Glu) and
K206E/K296E (in which both lysines are mutated to Glu), have been determined.
The K206E structure (2.6 A resolution; R = 0.213, R(free) = 0.269) shows that a
salt bridge is formed between Glu206 and Lys296, thus explaining the drastically
slower iron release by this mutant. The K206E/K296E double-mutant structure (2.8
A resolution; R = 0.232, R(free) = 0.259) shows that the Glu296 side chain moves
away from Glu206, easing any repulsive interaction and instead interacting with
the iron ligand His249. The evident conformational flexibility is consistent
with an alternative model for the operation of the dilysine pair in iron release
in which it facilitates concerted proton transfer to the tyrosine ligand Tyr188
as one step in the weakening of iron binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Dilysine interaction in the N-lobe of transferrin. (a)
Ribbon diagram showing the approach of Lys296 from domain N1
(lower) to Lys206 from domain N2 (upper) adjacent to the bound
Fe atom (red sphere). (b) Interactions made by the  -amino
group of Lys296: hydrogen bonds (broken lines) to Lys206 N^ -amino
group of Lys296: hydrogen bonds (broken lines) to Lys206 N^ 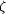 ,
Tyr188 O^ ,
Tyr188 O^ 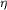 and
the aromatic ring of Tyr95. This and the other figures were
prepared using PyMOL (DeLano, 2002[DeLano, W. L. (2002). The
PyMOL Molecular Graphics System. http://www.pymol.org/ .]). and
the aromatic ring of Tyr95. This and the other figures were
prepared using PyMOL (DeLano, 2002[DeLano, W. L. (2002). The
PyMOL Molecular Graphics System. http://www.pymol.org/ .]).
|
 |
Figure 3.
Figure 3 Mutation site in the K206E/K296E double mutant. (a)
Bias-removed OMIT density (F[o] - F[c], contoured at 3  )
for the mutated residues. (b) Side chains of Glu206 and Glu296
in the mutant structure, shown in gold, superimposed on Lys206
and Lys296 in the wild-type structure, shown in silver. Other
residues common to both proteins are shown in green with atoms
coloured by type. Hydrogen bonds are shown as broken lines and
covalent metal-ligand bonds as solid black lines. )
for the mutated residues. (b) Side chains of Glu206 and Glu296
in the mutant structure, shown in gold, superimposed on Lys206
and Lys296 in the wild-type structure, shown in silver. Other
residues common to both proteins are shown in green with atoms
coloured by type. Hydrogen bonds are shown as broken lines and
covalent metal-ligand bonds as solid black lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2007,
63,
408-414)
copyright 2007.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.K.Khambati,
T.F.Moraes,
J.Singh,
S.R.Shouldice,
R.H.Yu,
and
A.B.Schryvers
(2010).
The role of vicinal tyrosine residues in the function of Haemophilus influenzae ferric-binding protein A.
|
| |
Biochem J,
432,
57-64.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

