 |
PDBsum entry 2jb3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2jb3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.2
- L-amino-acid oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an L-alpha-amino acid + O2 + H2O = a 2-oxocarboxylate + H2O2 + NH4+
|
 |
 |
 |
 |
 |
L-alpha-amino acid
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
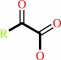 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
367:234-248
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of a bacterial L-amino acid oxidase from Rhodococcus opacus gives new evidence for the hydride mechanism for dehydrogenation.
|
|
A.Faust,
K.Niefind,
W.Hummel,
D.Schomburg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
l-Amino acid oxidase from Rhodococcus opacus (roLAAO) is classified as a member
of the GR(2)-family of flavin-dependent oxidoreductases according to a highly
conserved sequence motif for the cofactor binding. The monomer of the
homodimeric enzyme consists of three well-defined domains: the FAD-binding
domain corresponding to a general topology throughout the whole GR(2)-family; a
substrate-binding domain with almost the same topology as the snake venom LAAO
and a helical domain exclusively responsible for the unusual dimerisation mode
of the enzyme and not found in other members of the family so far. We describe
here high-resolution structures of the binary complex of protein and cofactor as
well as the ternary complexes of protein, cofactor and ligands. This structures
in addition to the structural knowledge of snake venom LAAO and DAAO from yeast
and pig kidney permit more insight into different steps in the reaction
mechanism of this class of enzymes. There is strong evidence for hydride
transfer as the mechanism of dehydrogenation. This mechanism appears to be
uncommon in a sense that the chemical transformation can proceed efficiently
without the involvement of amino acid functional groups. Most groups present at
the active site are involved in substrate recognition, binding and fixation,
i.e. they direct the trajectory of the interacting orbitals. In this mode of
catalysis orbital steering/interactions are the predominant factors for the
chemical step(s). A mirror-symmetrical relationship between the two
substrate-binding sites of d and l-amino acid oxidases is observed which
facilitates enantiomeric selectivity while preserving a common arrangement of
the residues in the active site. These results are of general relevance for the
mechanism of flavoproteins and lead to the proposal of a common dehydrogenation
step in the mechanism for l and d-amino acid oxidases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Schematic representation of the reaction catalyzed
by amino acid oxidases.
|
 |
Figure 10.
Figure 10. (a) Schematic representation of the Michaelis
complex M1. The interactions of the active site residues with
the substrate are shown by dotted lines. l-Alanine is bound in
the zwitterionic form. (b) Schematic representation of the
product complex P1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
367,
234-248)
copyright 2007.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.O.Baek,
J.W.Seo,
O.Kwon,
S.I.Seong,
I.H.Kim,
and
C.H.Kim
(2011).
Expression and characterization of a second L-amino acid deaminase isolated from Proteus mirabilis in Escherichia coli.
|
| |
J Basic Microbiol,
51,
129-135.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2010).
Oxidation of amines by flavoproteins.
|
| |
Arch Biochem Biophys,
493,
13-25.
|
 |
|
|
|
|
 |
M.H.Pozzi,
V.Gawandi,
and
P.F.Fitzpatrick
(2009).
Mechanistic studies of para-substituted N,N'-dibenzyl-1,4-diaminobutanes as substrates for a mammalian polyamine oxidase.
|
| |
Biochemistry,
48,
12305-12313.
|
 |
|
|
|
|
 |
S.Schriek,
U.Kahmann,
D.Staiger,
E.K.Pistorius,
and
K.P.Michel
(2009).
Detection of an L-amino acid dehydrogenase activity in Synechocystis sp. PCC 6803.
|
| |
J Exp Bot,
60,
1035-1046.
|
 |
|
|
|
|
 |
D.Georgieva,
A.Kardas,
F.Buck,
M.Perbandt,
and
C.Betzel
(2008).
Isolation, crystallization and preliminary X-ray diffraction analysis of L-amino-acid oxidase from Vipera ammodytes ammodytes venom.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
918-921.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

