 |
PDBsum entry 2isv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Structure of giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate
|
|
Structure:
|
 |
Putative fructose-1,6-bisphosphate aldolase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Giardia intestinalis. Organism_taxid: 5741. Strain: wb. Gene: ald. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.199
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
A.Galkin,O.Herzberg
|
Key ref:

|
 |
A.Galkin
et al.
(2007).
Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J Biol Chem,
282,
4859-4867.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-Oct-06
|
Release date:
|
12-Dec-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A8B2U2
(A8B2U2_GIAIC) -
Fructose-bisphosphate aldolase from Giardia intestinalis (strain ATCC 50803 / WB clone C6)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
323 a.a.
298 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
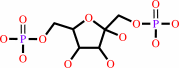 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:4859-4867
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
|
|
A.Galkin,
L.Kulakova,
E.Melamud,
L.Li,
C.Wu,
P.Mariano,
D.Dunaway-Mariano,
T.E.Nash,
O.Herzberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Class I and class II fructose-1,6-bisphosphate aldolases (FBPA), glycolytic
pathway enzymes, exhibit no amino acid sequence homology and utilize two
different catalytic mechanisms. The mammalian class I FBPA employs a Schiff base
mechanism, whereas the human parasitic protozoan Giardia lamblia class II FBPA
is a zinc-dependent enzyme. In this study, we have explored the potential
exploitation of the Giardia FBPA as a drug target. First, synthesis of FBPA was
demonstrated in Giardia trophozoites by using an antibody-based fluorescence
assay. Second, inhibition of FBPA gene transcription in Giardia trophozoites
suggested that the enzyme is necessary for the survival of the organism under
optimal laboratory growth conditions. Third, two crystal structures of FBPA in
complex with the transition state analog phosphoglycolohydroxamate (PGH) show
that the enzyme is homodimeric and that its active site contains a zinc ion. In
one crystal form, each subunit contains PGH, which is coordinated to the zinc
ion through the hydroxamic acid hydroxyl and carbonyl oxygen atoms. The second
crystal form contains PGH only in one subunit and the active site of the second
subunit is unoccupied. Inspection of the two states of the enzyme revealed that
it undergoes a conformational transition upon ligand binding. The enzyme cleaves
d-fructose-1,6-bisphosphate but not d-tagatose-1,6-bisphosphate, which is a
tight binding competitive inhibitor. The essential role of the active site
residue Asp-83 in catalysis was demonstrated by amino acid replacement.
Determinants of catalysis and substrate recognition, derived from comparison of
the G. lamblia FBPA structure with Escherichia coli FBPA and with a closely
related enzyme, E. coli tagatose-1,6-bisphosphate aldolase (TBPA), are described.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. A, reaction catalyzed by FBPA. B, FBPA inhibitor,
PGH, mimicking the DHAP ene-diolate transition-state
intermediate.
|
 |
Figure 2.
FIGURE 2. The catalytic mechanism of class I and class II
FBPA.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
4859-4867)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Li,
Z.Liu,
D.W.Cho,
J.Zou,
M.Gong,
R.M.Breece,
A.Galkin,
L.Li,
H.Zhao,
G.D.Maestas,
D.L.Tierney,
O.Herzberg,
D.Dunaway-Mariano,
and
P.S.Mariano
(2011).
Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
|
| |
J Inorg Biochem,
105,
509-517.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Galkin,
L.Kulakova,
R.Wu,
T.E.Nash,
D.Dunaway-Mariano,
and
O.Herzberg
(2010).
X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
386-390.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Wang,
H.Huang,
H.H.Nguyen,
K.N.Allen,
P.S.Mariano,
and
D.Dunaway-Mariano
(2010).
Divergence of biochemical function in the HAD superfamily: D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB).
|
| |
Biochemistry,
49,
1072-1081.
|
 |
|
|
|
|
 |
A.Galkin,
Z.Li,
L.Li,
L.Kulakova,
L.R.Pal,
D.Dunaway-Mariano,
and
O.Herzberg
(2009).
Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
48,
3186-3196.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.D.Pegan,
K.Rukseree,
S.G.Franzblau,
and
A.D.Mesecar
(2009).
Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
|
| |
J Mol Biol,
386,
1038-1053.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

