
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 179 a.a.
179 a.a.
|
 |

|

|

|

|

|

|

|
 182 a.a.
182 a.a.
|
 |

|

|

|

|

|

|

|
 15 a.a.
15 a.a.
|
 |

|

|

|

|

|

|

|
 198 a.a.
198 a.a.
|
 |

|

|

|

|

|

|

|
 239 a.a.
239 a.a.
|
 |

|

|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Immune system
|
 |
|
Title:
|
 |
Structural basis for recognition of mutant self by a tumor-specific, mhc class ii-restricted tcr
|
|
Structure:
|
 |
Hla class ii histocompatibility antigen, dr alpha chain. Chain: a. Fragment: residues 1-182 (26-207). Synonym: mhc class ii antigen dra, major histocompatibility complex class ii hla-dr1 alpha chain. Engineered: yes. Hla class ii histocompatibility antigen, drb1-1 beta chain. Chain: b. Fragment: residues 1-190 (30-219).
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: hla-dra. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693. Gene: hla-drb1. Gene: tpi1, tpi.
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.211
|
R-free:
|
0.279
|
|
|
Authors:
|
 |
L.Deng,R.J.Langley,R.A.Mariuzza
|
Key ref:

|
 |
L.Deng
et al.
(2007).
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor.
Nat Immunol,
8,
398-408.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Sep-06
|
Release date:
|
03-Apr-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P01903
(DRA_HUMAN) -
HLA class II histocompatibility antigen, DR alpha chain from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
254 a.a.
179 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P01911
(2B1F_HUMAN) -
HLA class II histocompatibility antigen, DRB1 beta chain from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
266 a.a.
182 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P60174
(TPIS_HUMAN) -
Triosephosphate isomerase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
249 a.a.
15 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chain P:
E.C.4.2.3.3
- methylglyoxal synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dihydroxyacetone phosphate = methylglyoxal + phosphate
|
 |
 |
 |
 |
 |
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
=
|
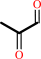 methylglyoxal
methylglyoxal
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain P:
E.C.5.3.1.1
- triose-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glyceraldehyde 3-phosphate = dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Immunol
8:398-408
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor.
|
|
L.Deng,
R.J.Langley,
P.H.Brown,
G.Xu,
L.Teng,
Q.Wang,
M.I.Gonzales,
G.G.Callender,
M.I.Nishimura,
S.L.Topalian,
R.A.Mariuzza.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Structural studies of complexes of T cell receptor (TCR) and peptide-major
histocompatibility complex (MHC) have focused on TCRs specific for foreign
antigens or native self. An unexplored category of TCRs includes those specific
for self determinants bearing alterations resulting from disease, notably
cancer. We determined here the structure of a human melanoma-specific TCR (E8)
bound to the MHC molecule HLA-DR1 and an epitope from mutant triosephosphate
isomerase. The structure had features intermediate between 'anti-foreign' and
autoimmune TCR-peptide-MHC class II complexes that may reflect the hybrid nature
of altered self. E8 manifested very low affinity for mutant triosephosphate
isomerase-HLA-DR1 despite the highly tumor-reactive properties of E8 cells. A
second TCR (G4) had even lower affinity but underwent peptide-specific formation
of dimers, suggesting this as a mechanism for enhancing low-affinity
TCR-peptide-MHC interactions for T cell activation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
(a) TCR residues in contact with mutant TPI. The substituted
TPI isoleucine residue (position 3) is magenta. (b) Interactions
between E8 and mutant TPI. (c) Interactions between the E8 V[
 ]domain
and the HLA-DR1 ]domain
and the HLA-DR1  1 1
 -helix.
(d,e) Interactions between mutant TPI isoleucine at position 3
and TCR E8 (d) or wild-type TPI threonine at position 3 and
HLA-DR1 complexes (e). TCR residues are identified by one-letter
amino-acid designation followed by position number and chain
designation; peptide residues are identified by one-letter
amino-acid designation followed by position ('P') number. Colors
of TCR and MHC molecules as in Figure 3; hydrogen bonds, red
dotted lines; salt bridges, solid lines; van der Waals contacts,
black dotted lines. -helix.
(d,e) Interactions between mutant TPI isoleucine at position 3
and TCR E8 (d) or wild-type TPI threonine at position 3 and
HLA-DR1 complexes (e). TCR residues are identified by one-letter
amino-acid designation followed by position number and chain
designation; peptide residues are identified by one-letter
amino-acid designation followed by position ('P') number. Colors
of TCR and MHC molecules as in Figure 3; hydrogen bonds, red
dotted lines; salt bridges, solid lines; van der Waals contacts,
black dotted lines.
|
 |
Figure 6.
(a–c) Structural rearrangements in CDR2  ,
CDR3 ,
CDR3  and
CDR3 and
CDR3  induced
by binding to mutant TPI–DR1. Bound E8, green; unbound E8,
magenta; HLA-DR1, gold; mutant TPI, gray. Residues from E8 and
DR1 involved in interactions with peptide are identified. (d)
Superposition of mutant TPI–DR1 in a form without bound ligand
(peptide, cyan; MHC, violet) and in complex with E8 (peptide,
gray; MHC, gold). Arrow indicates residues 55–59 of the
HLA-DR1 induced
by binding to mutant TPI–DR1. Bound E8, green; unbound E8,
magenta; HLA-DR1, gold; mutant TPI, gray. Residues from E8 and
DR1 involved in interactions with peptide are identified. (d)
Superposition of mutant TPI–DR1 in a form without bound ligand
(peptide, cyan; MHC, violet) and in complex with E8 (peptide,
gray; MHC, gold). Arrow indicates residues 55–59 of the
HLA-DR1  1
domain. (e) Conformational changes in TPI residues that contact
E8. Residues of TPI involved in interactions with E8 are
identified. Residue designations as in Figure 4. 1
domain. (e) Conformational changes in TPI residues that contact
E8. Residues of TPI involved in interactions with E8 are
identified. Residue designations as in Figure 4.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Immunol
(2007,
8,
398-408)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Khan,
and
S.Ranganathan
(2011).
Understanding TR Binding to pMHC Complexes: How Does a TR Scan Many pMHC Complexes yet Preferentially Bind to One.
|
| |
PLoS One,
6,
e17194.
|
 |
|
|
|
|
 |
P.Schuck
(2010).
On computational approaches for size-and-shape distributions from sedimentation velocity analytical ultracentrifugation.
|
| |
Eur Biophys J,
39,
1261-1275.
|
 |
|
|
|
|
 |
S.P.Persaud,
D.L.Donermeyer,
K.S.Weber,
D.M.Kranz,
and
P.M.Allen
(2010).
High-affinity T cell receptor differentiates cognate peptide-MHC and altered peptide ligands with distinct kinetics and thermodynamics.
|
| |
Mol Immunol,
47,
1793-1801.
|
 |
|
|
|
|
 |
G.Chen,
G.Han,
J.Feng,
J.Wang,
R.Wang,
R.Xu,
B.Shen,
J.Qian,
and
Y.Li
(2009).
Glutamic acid decarboxylase-derived epitopes with specific domains expand CD4(+)CD25(+) regulatory T cells.
|
| |
PLoS One,
4,
e7034.
|
 |
|
|
|
|
 |
K.W.Wucherpfennig,
M.J.Call,
L.Deng,
and
R.Mariuzza
(2009).
Structural alterations in peptide-MHC recognition by self-reactive T cell receptors.
|
| |
Curr Opin Immunol,
21,
590-595.
|
 |
|
|
|
|
 |
M.Harkiolaki,
S.L.Holmes,
P.Svendsen,
J.W.Gregersen,
L.T.Jensen,
R.McMahon,
M.A.Friese,
G.van Boxel,
R.Etzensperger,
J.S.Tzartos,
K.Kranc,
S.Sainsbury,
K.Harlos,
E.D.Mellins,
J.Palace,
M.M.Esiri,
P.A.van der Merwe,
E.Y.Jones,
and
L.Fugger
(2009).
T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides.
|
| |
Immunity,
30,
348-357.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Beddoe,
Z.Chen,
C.S.Clements,
L.K.Ely,
S.R.Bushell,
J.P.Vivian,
L.Kjer-Nielsen,
S.S.Pang,
M.A.Dunstone,
Y.C.Liu,
W.A.Macdonald,
M.A.Perugini,
M.C.Wilce,
S.R.Burrows,
A.W.Purcell,
T.Tiganis,
S.P.Bottomley,
J.McCluskey,
and
J.Rossjohn
(2009).
Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor.
|
| |
Immunity,
30,
777-788.
|
 |
|
|
|
|
 |
D.I.Godfrey,
J.Rossjohn,
and
J.McCluskey
(2008).
The fidelity, occasional promiscuity, and versatility of T cell receptor recognition.
|
| |
Immunity,
28,
304-314.
|
 |
|
|
|
|
 |
K.M.Armstrong,
K.H.Piepenbrink,
and
B.M.Baker
(2008).
Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes.
|
| |
Biochem J,
415,
183-196.
|
 |
|
|
|
|
 |
P.Marrack,
J.P.Scott-Browne,
S.Dai,
L.Gapin,
and
J.W.Kappler
(2008).
Evolutionarily conserved amino acids that control TCR-MHC interaction.
|
| |
Annu Rev Immunol,
26,
171-203.
|
 |
|
|
|
|
 |
P.Marrack,
K.Rubtsova,
J.Scott-Browne,
and
J.W.Kappler
(2008).
T cell receptor specificity for major histocompatibility complex proteins.
|
| |
Curr Opin Immunol,
20,
203-207.
|
 |
|
|
|
|
 |
C.Mazza,
and
B.Malissen
(2007).
What guides MHC-restricted TCR recognition?
|
| |
Semin Immunol,
19,
225-235.
|
 |
|
|
|
|
 |
L.Deng,
and
R.A.Mariuzza
(2007).
Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors.
|
| |
Trends Biochem Sci,
32,
500-508.
|
 |
|
|
|
|
 |
M.E.Call,
and
K.W.Wucherpfennig
(2007).
Common themes in the assembly and architecture of activating immune receptors.
|
| |
Nat Rev Immunol,
7,
841-850.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|

| |

