 |
PDBsum entry 2htn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal binding protein, oxidoreductase
|
PDB id
|
|

|

|
2htn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Metal binding protein, oxidoreductase
|
 |
|
Title:
|
 |
E. Coli bacterioferritin in its as-isolated form
|
|
Structure:
|
 |
Bacterioferritin. Chain: a, b, c, d, e, f, g, h. Synonym: bfr, cytochrome b-1, cytochrome b-557. Ec: 1.16.3.1
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 511693. Strain: bl21
|
|
Biol. unit:
|
 |
 24mer (from PDB file)
24mer (from PDB file)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.172
|
R-free:
|
0.188
|
|
|
Authors:
|
 |
A.Van Eerde,S.Wolterink-Van Loo,J.Van Der Oost,B.W.Dijkstra
|
Key ref:

|
 |
A.van Eerde
et al.
(2006).
Fortuitous structure determination of 'as-isolated' Escherichia coli bacterioferritin in a novel crystal form.
Acta Crystallograph Sect F Struct Biol Cryst Commun,
62,
1061-1066.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Jul-06
|
Release date:
|
21-Nov-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0ABD3
(BFR_ECOLI) -
Bacterioferritin from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
158 a.a.
158 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.16.3.1
- ferroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 Fe2+ + O2 + 4 H+ = 4 Fe3+ + 2 H2O
|
 |
 |
 |
 |
 |
4
×
Fe(2+)
|
+
|
 O2
O2
|
+
|
4
×
H(+)
|
=
|
4
×
Fe(3+)
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallograph Sect F Struct Biol Cryst Commun
62:1061-1066
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Fortuitous structure determination of 'as-isolated' Escherichia coli bacterioferritin in a novel crystal form.
|
|
A.van Eerde,
S.Wolterink-van Loo,
J.van der Oost,
B.W.Dijkstra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Escherichia coli bacterioferritin was serendipitously crystallized in a novel
cubic crystal form and its structure could be determined to 2.5 A resolution
despite a high degree of merohedral twinning. This is the first report of
crystallographic data on 'as-isolated' E. coli bacterioferritin. The ferroxidase
active site contains positive difference density consistent with two metal ions
that had co-purified with the protein. X-ray fluorescence studies suggest that
the metal composition is different from that of previous structures and is a mix
of zinc and native iron ions. The ferroxidase-centre configuration displays a
similar flexibility as previously noted for other bacterioferritins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 The ferroxidase centre. (a) Stereo figures of  [A]-weighted
F[o] - F[c] density in the ferroxidase site contoured at 5 [A]-weighted
F[o] - F[c] density in the ferroxidase site contoured at 5  ,
(b) superposition of the ferroxidase centre of the P2[1]3
structure (grey/orange) and the Mn-bound structure
(yellow/purple; PDB code 1bcf ; Frolow et al., 1994[Frolow, F.,
Kalb, A. J. & Yariv, J. (1994). Nature Struct. Biol. 1,
453-460.]) of E. coli bacterioferritin and (c) superposition of
the FE2 site environment in the P2[1]3 E. coli bacterioferritin
structure (grey) with the corresponding region in the structure
of reduced A. vinelandii bacterioferritin (pale green; PDB code
1fkz ; Swartz et al., 2006[Swartz, L., Kuchinskas, M., Li, H.,
Poulos, T. L. & Lanzilotta, W. N. (2006). Biochemistry, 45,
4421-4428.]). The location of the inner cavity (IC) is
indicated. This view is rotated ,
(b) superposition of the ferroxidase centre of the P2[1]3
structure (grey/orange) and the Mn-bound structure
(yellow/purple; PDB code 1bcf ; Frolow et al., 1994[Frolow, F.,
Kalb, A. J. & Yariv, J. (1994). Nature Struct. Biol. 1,
453-460.]) of E. coli bacterioferritin and (c) superposition of
the FE2 site environment in the P2[1]3 E. coli bacterioferritin
structure (grey) with the corresponding region in the structure
of reduced A. vinelandii bacterioferritin (pale green; PDB code
1fkz ; Swartz et al., 2006[Swartz, L., Kuchinskas, M., Li, H.,
Poulos, T. L. & Lanzilotta, W. N. (2006). Biochemistry, 45,
4421-4428.]). The location of the inner cavity (IC) is
indicated. This view is rotated 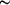 90°
with respect to the previous views. 90°
with respect to the previous views.
|
 |
Figure 4.
Figure 4 X-ray fluorescence scans around the K edges of Mn, Fe,
Cu and Zn, as indicated. Plots are corrected for beam intensity
and fluorescence is on an arbitrary scale.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the IUCr:
Acta Crystallograph Sect F Struct Biol Cryst Commun
(2006,
62,
1061-1066)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.G.Wong,
S.A.Tom-Yew,
A.Lewin,
N.E.Le Brun,
G.R.Moore,
M.E.Murphy,
and
A.G.Mauk
(2009).
Structural and Mechanistic Studies of a Stabilized Subunit Dimer Variant of Escherichia coli Bacterioferritin Identify Residues Required for Core Formation.
|
| |
J Biol Chem,
284,
18873-18881.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Janowski,
T.Auerbach-Nevo,
and
M.S.Weiss
(2008).
Bacterioferritin from Mycobacterium smegmatis contains zinc in its di-nuclear site.
|
| |
Protein Sci,
17,
1138-1150.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Gupta,
R.K.Gupta,
G.Khare,
D.M.Salunke,
and
A.K.Tyagi
(2008).
Cloning, expression, purification, crystallization and preliminary X-ray crystallographic analysis of bacterioferritin A from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
398-401.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

