
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the naphthalene 1,2-dioxygenase f352v mutant bound to phenanthrene.
|
|
Structure:
|
 |
Naphthalene 1,2-dioxygenase alpha subunit. Chain: a. Synonym: naphthalene 1,2-dioxygenase isp alpha. Engineered: yes. Mutation: yes. Naphthalene 1,2-dioxygenase beta subunit. Chain: b. Synonym: naphthalene 1,2-dioxygenase isp beta. Engineered: yes
|
|
Source:
|
 |
Pseudomonas sp.. Organism_taxid: 306. Gene: doxb. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: doxd.
|
|
Biol. unit:
|
 |
 Hexamer (from PDB file)
Hexamer (from PDB file)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.165
|
R-free:
|
0.190
|
|
|
Authors:
|
 |
D.J.Ferraro,A.L.Okerlund,J.C.Mowers,S.Ramaswamy
|
|
Key ref:
|
 |
D.J.Ferraro
et al.
(2006).
Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J Bacteriol,
188,
6986-6994.
PubMed id:
|
 |
|
Date:
|
 |
|
11-Jul-06
|
Release date:
|
10-Oct-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.1.14.12.12
- naphthalene 1,2-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Aromatic 1,2-Dioxygenases
|
 |
 |
 |
 |
 |
Reaction:
|
 |
naphthalene + NADH + O2 + H+ = (1R,2S)-1,2-dihydronaphthalene-1,2-diol + NAD+
|
 |
 |
 |
 |
 |
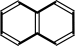 naphthalene
naphthalene
|
+
|
 NADH
NADH
|
+
|
O2
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
+
|
H(+)
|
=
|
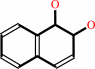 (1R,2S)-1,2-dihydronaphthalene-1,2-diol
(1R,2S)-1,2-dihydronaphthalene-1,2-diol
|
+
|
 NAD(+)
NAD(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
188:6986-6994
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
|
|
D.J.Ferraro,
A.L.Okerlund,
J.C.Mowers,
S.Ramaswamy.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Rieske oxygenase (RO) systems are two- and three-component enzyme systems that
catalyze the formation of cis-dihydrodiols from aromatic substrates. Degradation
of pollutants in contaminated soil and generation of chiral synthons have been
the major foci of RO research. Substrate specificity and product regio- and
stereoselectivity have been shown to vary between individual ROs. While directed
evolution methods for altering RO function have been successful in the past,
rational engineering of these enzymes still poses a challenge due to the lack of
structural understanding. Here we examine the structural changes induced by
mutation of Phe-352 in naphthalene 1,2-dioxygenase from Pseudomonas sp. strain
NCIB 9816-4 (NDO-O(9816-4)). Structures of the Phe-352-Val mutant in native form
and in complex with phenanthrene and anthracene, along with those of wild-type
NDO-O(9816-4) in complex with phenanthrene, anthracene, and 3-nitrotoluene, are
presented. Phenanthrene was shown to bind in a different orientation in the
Phe-352-Val mutant active site from that in the wild type, while anthracene was
found to bind in similar positions in both enzymes. Two orientations of
3-nitrotoluene were observed, i.e., a productive and a nonproductive
orientation. These orientations help explain why NDO-O(9816-4) forms different
products from 3-nitrotoluene than those made from nitrobenzene dioxygenase.
Comparison of these structures among themselves and with other known ROs bound
to substrates reveals that the orientation of substrate binding at the active
site is the primary determinant of product regio- and stereoselectivity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Mallick,
J.Chakraborty,
and
T.K.Dutta
(2011).
Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: a review.
|
| |
Crit Rev Microbiol,
37,
64-90.
|
 |
|
|
|
|
 |
J.Seo,
S.I.Kang,
J.Y.Ryu,
Y.J.Lee,
K.D.Park,
M.Kim,
D.Won,
H.Y.Park,
J.H.Ahn,
Y.Chong,
R.A.Kanaly,
J.Han,
and
H.G.Hur
(2010).
Location of flavone B-ring controls regioselectivity and stereoselectivity of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4.
|
| |
Appl Microbiol Biotechnol,
86,
1451-1462.
|
 |
|
|
|
|
 |
O.Kweon,
S.J.Kim,
J.P.Freeman,
J.Song,
S.Baek,
and
C.E.Cerniglia
(2010).
Substrate Specificity and Structural Characteristics of the Novel Rieske Nonheme Iron Aromatic Ring-Hydroxylating Oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1.
|
| |
MBio,
1,
0.
|
 |
|
|
|
|
 |
D.R.Boyd,
N.D.Sharma,
G.P.Coen,
F.Hempenstall,
V.Ljubez,
J.F.Malone,
C.C.Allen,
and
J.T.Hamilton
(2008).
Regioselectivity and stereoselectivity of dioxygenase catalysed cis-dihydroxylation of mono- and tri-cyclic azaarene substrates.
|
| |
Org Biomol Chem,
6,
3957-3966.
|
 |
|
|
|
|
 |
H.Uchimura,
T.Horisaki,
T.Umeda,
H.Noguchi,
Y.Usami,
L.Li,
T.Terada,
S.Nakamura,
K.Shimizu,
T.Takemura,
H.Habe,
K.Furihata,
T.Omori,
H.Yamane,
and
H.Nojiri
(2008).
Alteration of the substrate specificity of the angular dioxygenase carbazole 1,9a-dioxygenase.
|
| |
Biosci Biotechnol Biochem,
72,
3237-3248.
|
 |
|
|
|
|
 |
P.C.Bruijnincx,
G.van Koten,
and
R.J.Klein Gebbink
(2008).
Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies.
|
| |
Chem Soc Rev,
37,
2716-2744.
|
 |
|
|
|
|
 |
T.D.Bugg,
and
S.Ramaswamy
(2008).
Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations.
|
| |
Curr Opin Chem Biol,
12,
134-140.
|
 |
|
|
|
|
 |
T.Ohta,
S.Chakrabarty,
J.D.Lipscomb,
and
E.I.Solomon
(2008).
Near-IR MCD of the nonheme ferrous active site in naphthalene 1,2-dioxygenase: correlation to crystallography and structural insight into the mechanism of Rieske dioxygenases.
|
| |
J Am Chem Soc,
130,
1601-1610.
|
 |
|
|
|
|
 |
D.J.Ferraro,
E.N.Brown,
C.L.Yu,
R.E.Parales,
D.T.Gibson,
and
S.Ramaswamy
(2007).
Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
|
| |
BMC Struct Biol,
7,
10.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

