 |
PDBsum entry 2gps

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.6.3.4.14
- biotin carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-biotinyl-L-lysyl-[protein] + hydrogencarbonate + ATP = N6- carboxybiotinyl-L-lysyl-[protein] + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
N(6)-biotinyl-L-lysyl-[protein]
|
+
|
 hydrogencarbonate
hydrogencarbonate
|
+
|
 ATP
ATP
|
=
|
N(6)- carboxybiotinyl-L-lysyl-[protein]
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.4.1.2
- acetyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + acetyl-CoA + ATP = malonyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 hydrogencarbonate
hydrogencarbonate
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
 ATP
ATP
|
=
|
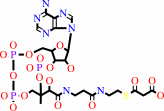 malonyl-CoA
malonyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
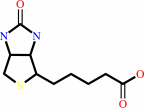 Biotin
Biotin
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Cell
22:807-818
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
|
|
Y.Shen,
C.Y.Chou,
G.G.Chang,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acetyl-coenzyme A carboxylases (ACCs) have crucial roles in fatty acid
metabolism. The biotin carboxylase (BC) subunit of Escherichia coli ACC is
believed to be active only as a dimer, although the crystal structure shows that
the active site of each monomer is 25 A from the dimer interface. We report here
biochemical, biophysical, and structural characterizations of BC carrying
single-site mutations in the dimer interface. Our studies demonstrate that two
of the mutants, R19E and E23R, are monomeric in solution but have only a 3-fold
loss in catalytic activity. The crystal structures of the E23R and F363A mutants
show that they can still form the correct dimer at high concentrations. Our data
suggest that dimerization is not an absolute requirement for the catalytic
activity of the E. coli BC subunit, and we propose a new model for the molecular
mechanism of action for BC in multisubunit and multidomain ACCs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Sedimentation Velocity Analytical
Ultracentrifugation Data for the BC Subunit
|
 |
Figure 5.
Figure 5. A Model for the Mechanism of Action of BC
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2006,
22,
807-818)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.R.Novak,
D.Moldovan,
G.L.Waldrop,
and
M.S.de Queiroz
(2011).
Behavior of the ATP grasp domain of biotin carboxylase monomers and dimers studied using molecular dynamics simulations.
|
| |
Proteins,
79,
622-632.
|
 |
|
|
|
|
 |
E.F.Franca,
F.L.Leite,
R.A.Cunha,
O.N.Oliveira,
and
L.C.Freitas
(2011).
Designing an enzyme-based nanobiosensor using molecular modeling techniques.
|
| |
Phys Chem Chem Phys,
13,
8894-8899.
|
 |
|
|
|
|
 |
C.S.Huang,
K.Sadre-Bazzaz,
Y.Shen,
B.Deng,
Z.H.Zhou,
and
L.Tong
(2010).
Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
|
| |
Nature,
466,
1001-1005.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.C.Cheng,
G.G.Chang,
and
C.Y.Chou
(2010).
Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease.
|
| |
Biophys J,
98,
1327-1336.
|
 |
|
|
|
|
 |
C.Y.Chou,
L.P.Yu,
and
L.Tong
(2009).
Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
|
| |
J Biol Chem,
284,
11690-11697.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.P.Yu,
S.Xiang,
G.Lasso,
D.Gil,
M.Valle,
and
L.Tong
(2009).
A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A.
|
| |
Structure,
17,
823-832.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Mochalkin,
J.R.Miller,
A.Evdokimov,
S.Lightle,
C.Yan,
C.K.Stover,
and
G.L.Waldrop
(2008).
Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase.
|
| |
Protein Sci,
17,
1706-1718.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Jitrapakdee,
K.H.Surinya,
A.Adina-Zada,
S.W.Polyak,
C.Stojkoski,
R.Smyth,
G.W.Booker,
W.W.Cleland,
P.V.Attwood,
and
J.C.Wallace
(2007).
Conserved Glu40 and Glu433 of the biotin carboxylase domain of yeast pyruvate carboxylase I isoenzyme are essential for the association of tetramers.
|
| |
Int J Biochem Cell Biol,
39,
2120-2134.
|
 |
|
|
|
|
 |
L.Tong,
and
H.J.Harwood
(2006).
Acetyl-coenzyme A carboxylases: versatile targets for drug discovery.
|
| |
J Cell Biochem,
99,
1476-1488.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

