 |
PDBsum entry 2gfq

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
2gfq
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Structure of protein of unknown function ph0006 from pyrococcus horikoshii
|
|
Structure:
|
 |
Upf0204 protein ph0006. Chain: a, b, c. Engineered: yes
|
|
Source:
|
 |
Pyrococcus horikoshii. Organism_taxid: 70601. Strain: ot3. Gene: ph0006. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.75Å
|
R-factor:
|
0.152
|
R-free:
|
0.179
|
|
|
Authors:
|
 |
M.E.Cuff,T.Skarina,E.Gorodichtchenskaia,A.Edwards,A.Savchenko, A.Joachimiak,Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
M.E.Cuff
et al.
Structure of hypothetical protein ph0006 from pyrococcus horikoshii.
To be published,
.
|
 |
|
Date:
|
 |
|
22-Mar-06
|
Release date:
|
25-Apr-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O57774
(DTDA_PYRHO) -
D-aminoacyl-tRNA deacylase from Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
274 a.a.
288 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.96
- D-aminoacyl-tRNA deacylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a D-aminoacyl-tRNA + H2O = a tRNA + a D-alpha-amino acid + H+
|
|
2.
|
glycyl-tRNA(Ala) + H2O = tRNA(Ala) + glycine + H+
|
|
 |
 |
 |
 |
 |
D-aminoacyl-tRNA
|
+
|
H2O
|
=
|
 tRNA
tRNA
|
+
|
D-alpha-amino acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
glycyl-tRNA(Ala)
|
+
|
H2O
|
=
|
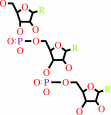 tRNA(Ala)
tRNA(Ala)
|
+
|
 glycine
glycine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

