 |
PDBsum entry 2g2n

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Unknown function
|
PDB id
|
|

|

|
2g2n
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.2.17
- hydroxyisourate hydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-hydroxyisourate + H2O = 5-hydroxy-2-oxo-4-ureido-2,5-dihydro- 1H-imidazole-5-carboxylate + H+
|
 |
 |
 |
 |
 |
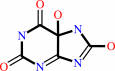 5-hydroxyisourate
5-hydroxyisourate
|
+
|
H2O
|
=
|
5-hydroxy-2-oxo-4-ureido-2,5-dihydro- 1H-imidazole-5-carboxylate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Struct Biol
155:445-457
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The transthyretin-related protein: structural investigation of a novel protein family.
|
|
E.Lundberg,
S.Bäckström,
U.H.Sauer,
A.E.Sauer-Eriksson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The transthyretin-related protein (TRP) family comprises proteins predicted to
be structurally related to the homotetrameric transport protein transthyretin
(TTR). The function of TRPs is not yet fully established, but recent data
suggest that they are involved in purine catabolism. We have determined the
three-dimensional structure of the Escherichia coli TRP in two crystal forms;
one at 1.65 A resolution in the presence of zinc, and the other at 2.1 A
resolution in the presence of zinc and bromide. The structures revealed five
zinc-ion-binding sites per monomer. Of these, the zinc ions bound at sites I and
II are coordinated in tetrahedral geometries to the side chains of residues
His9, His96, His98, Ser114, and three water molecules at the putative
ligand-binding site. Of these four residues, His9, His98, and Ser114 are
conserved. His9 and His98 bind the central zinc (site I) together with two water
molecules. The side chain of His98 also binds to the zinc ion at site II.
Bromide ions bind at site I only, replacing one of the water molecules
coordinated to the zinc ion. The C-terminal four amino acid sequence motif
Y-[RK]-G-[ST] constitutes the signature sequence of the TRP family. Two Tyr111
residues form direct hydrogen bonds to each other over the tetramer interface at
the area, which in TTR constitutes the rear part of its thyroxine-binding
channel. The putative substrate/ligand-binding channel of TRP is consequently
shallower and broader than its counterpart in TTR.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Pessoa,
Z.Sárkány,
F.Ferreira-da-Silva,
S.Martins,
M.R.Almeida,
J.Li,
and
A.M.Damas
(2010).
Functional characterization of Arabidopsis thaliana transthyretin-like protein.
|
| |
BMC Plant Biol,
10,
30.
|
 |
|
|
|
|
 |
X.Wang,
W.Li,
D.Zhao,
B.Liu,
Y.Shi,
B.Chen,
H.Yang,
P.Guo,
X.Geng,
Z.Shang,
E.Peden,
E.Kage-Nakadai,
S.Mitani,
and
D.Xue
(2010).
Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor.
|
| |
Nat Cell Biol,
12,
655-664.
|
 |
|
|
|
|
 |
E.Lundberg,
A.Olofsson,
G.T.Westermark,
and
A.E.Sauer-Eriksson
(2009).
Stability and fibril formation properties of human and fish transthyretin, and of the Escherichia coli transthyretin-related protein.
|
| |
FEBS J,
276,
1999-2011.
|
 |
|
|
|
|
 |
S.C.Hennebry
(2009).
Evolutionary changes to transthyretin: structure and function of a transthyretin-like ancestral protein.
|
| |
FEBS J,
276,
5367-5379.
|
 |
|
|
|
|
 |
G.Zanotti,
C.Folli,
L.Cendron,
B.Alfieri,
S.K.Nishida,
F.Gliubich,
N.Pasquato,
A.Negro,
and
R.Berni
(2008).
Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
|
| |
FEBS J,
275,
5841-5854.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Kim,
J.Park,
and
S.Rhee
(2007).
Structural and functional basis for (S)-allantoin formation in the ureide pathway.
|
| |
J Biol Chem,
282,
23457-23464.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Cendron,
R.Berni,
C.Folli,
I.Ramazzina,
R.Percudani,
and
G.Zanotti
(2007).
The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
|
| |
J Biol Chem,
282,
18182-18189.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

