 |
PDBsum entry 2fr5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.5
- cytidine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
cytidine + H2O + H+ = uridine + NH4+
|
|
2.
|
2'-deoxycytidine + H2O + H+ = 2'-deoxyuridine + NH4+
|
|
 |
 |
 |
 |
 |
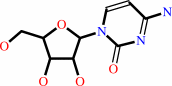 cytidine
cytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
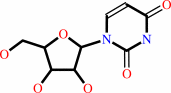 uridine
uridine
|
+
|
NH4(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 2'-deoxycytidine
2'-deoxycytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyuridine
Bound ligand (Het Group name = )
matches with 94.12% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
45:7825-7833
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The 1.48 A resolution crystal structure of the homotetrameric cytidine deaminase from mouse.
|
|
A.H.Teh,
M.Kimura,
M.Yamamoto,
N.Tanaka,
I.Yamaguchi,
T.Kumasaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytidine deaminase (CDA) is a zinc-dependent enzyme that catalyzes the
deamination of cytidine or deoxycytidine to form uridine or deoxyuridine. Here
we present the crystal structure of mouse CDA (MmCDA), complexed with either
tetrahydrouridine (THU), 3-deazauridine (DAU), or cytidine. In the MmCDA-DAU
complex, it clearly demonstrates that cytidine is distinguished from uridine by
its 4-NH(2) group that acts as a hydrogen bond donor. In the MmCDA-cytidine
complex, cytidine, unexpectedly, binds as the substrate instead of the
deaminated product in three of the four subunits, and in the remaining subunit
it binds as the product uridine. Furthermore, the charge-neutralizing Arg68 of
MmCDA has also exhibited two alternate conformations, I and II. In conformation
I, the only conformation observed in the other structurally known homotetrameric
CDAs, Arg68 hydrogen bonds Cys65 and Cys102 to modulate part of their negative
charges. However, in conformation II the side chain of Arg68 rotates about 130
degrees around the Cgamma-Cdelta bond and abolishes these hydrogen bonds. The
lack of hydrogen bonding may indirectly weaken the zinc-product interaction by
increased electron donation from cysteine to the zinc ion, suggesting a novel
product-expelling mechanism. On the basis of known structures, structural
analysis further reveals two subclasses of homotetrameric CDAs that can be
identified according to the position of the charge-neutralizing arginine
residue. Implications for CDA-RNA interaction have also been considered.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.A.Sánchez-Quitian,
L.F.Timmers,
R.A.Caceres,
J.G.Rehm,
C.E.Thompson,
L.A.Basso,
W.F.de Azevedo,
and
D.S.Santos
(2011).
Crystal structure determination and dynamic studies of Mycobacterium tuberculosis Cytidine deaminase in complex with products.
|
| |
Arch Biochem Biophys,
509,
108-115.
|
 |
|
|
|
|
 |
A.Furukawa,
T.Nagata,
A.Matsugami,
Y.Habu,
R.Sugiyama,
F.Hayashi,
N.Kobayashi,
S.Yokoyama,
H.Takaku,
and
M.Katahira
(2009).
Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G.
|
| |
EMBO J,
28,
440-451.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.R.Ludek,
G.K.Schroeder,
C.Liao,
P.L.Russ,
R.Wolfenden,
and
V.E.Marquez
(2009).
Synthesis and conformational analysis of locked carbocyclic analogues of 1,3-diazepinone riboside, a high-affinity cytidine deaminase inhibitor.
|
| |
J Org Chem,
74,
6212-6223.
|
 |
|
|
|
|
 |
V.E.Marquez,
G.K.Schroeder,
O.R.Ludek,
M.A.Siddiqui,
A.Ezzitouni,
and
R.Wolfenden
(2009).
Contrasting behavior of conformationally locked carbocyclic nucleosides of adenosine and cytidine as substrates for deaminases.
|
| |
Nucleosides Nucleotides Nucleic Acids,
28,
614-632.
|
 |
|
|
|
|
 |
X.Xu,
H.C.Hsu,
J.Chen,
W.E.Grizzle,
W.W.Chatham,
C.R.Stockard,
Q.Wu,
P.A.Yang,
V.M.Holers,
and
J.D.Mountz
(2009).
Increased expression of activation-induced cytidine deaminase is associated with anti-CCP and rheumatoid factor in rheumatoid arthritis.
|
| |
Scand J Immunol,
70,
309-316.
|
 |
|
|
|
|
 |
K.M.Chen,
E.Harjes,
P.J.Gross,
A.Fahmy,
Y.Lu,
K.Shindo,
R.S.Harris,
and
H.Matsuo
(2008).
Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
|
| |
Nature,
452,
116-119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Prochnow,
R.Bransteitter,
M.G.Klein,
M.F.Goodman,
and
X.S.Chen
(2007).
The APOBEC-2 crystal structure and functional implications for the deaminase AID.
|
| |
Nature,
445,
447-451.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kumasaka,
M.Yamamoto,
M.Furuichi,
M.Nakasako,
A.H.Teh,
M.Kimura,
I.Yamaguchi,
and
T.Ueki
(2007).
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
|
| |
J Biol Chem,
282,
37103-37111.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

