 |
PDBsum entry 2fit

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Chromosomal translocation
|
PDB id
|
|

|

|
2fit
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.51
- adenylylsulfate--ammonia adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
adenosine 5'-phosphosulfate + NH4+ = adenosine 5'-phosphoramidate + sulfate + 2 H+
|
 |
 |
 |
 |
 |
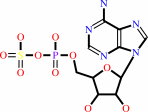 adenosine 5'-phosphosulfate
adenosine 5'-phosphosulfate
|
+
|
NH4(+)
|
=
|
 adenosine 5'-phosphoramidate
adenosine 5'-phosphoramidate
|
+
|
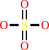 sulfate
sulfate
|
+
|
2
×
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.6.1.29
- bis(5'-adenosyl)-triphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
P1,P3-bis(5'-adenosyl) triphosphate + H2O = AMP + ADP + 2 H+
|
 |
 |
 |
 |
 |
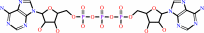 P(1),P(3)-bis(5'-adenosyl) triphosphate
P(1),P(3)-bis(5'-adenosyl) triphosphate
|
+
|
H2O
|
=
|
 AMP
AMP
|
+
|
 ADP
ADP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.6.2.1
- adenylylsulfatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
adenosine 5'-phosphosulfate + H2O = sulfate + AMP + 2 H+
|
 |
 |
 |
 |
 |
 adenosine 5'-phosphosulfate
adenosine 5'-phosphosulfate
|
+
|
H2O
|
=
|
 sulfate
sulfate
|
+
|
 AMP
AMP
|
+
|
2
×
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.3.9.1.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
5:763-774
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
|
|
C.D.Lima,
K.L.D'Amico,
I.Naday,
G.Rosenbaum,
E.M.Westbrook,
W.A.Hendrickson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The fragile histidine triad (FHIT) protein is a member of the large
and ubiquitous histidine triad (HIT) family of proteins. It is expressed from a
gene located at a fragile site on human chromosome 3, which is commonly
disrupted in association with certain cancers. On the basis of the genetic
evidence, it has been postulated that the FHIT protein may function as a tumor
suppressor, implying a role for the FHIT protein in carcinogenesis. The FHIT
protein has dinucleoside polyphosphate hydrolase activity in vitro, thus
suggesting that its role in vivo may involve the hydrolysis of a
phosphoanhydride bond. The structural analysis of FHIT will identify critical
residues involved in substrate binding and catalysis, and will provide insights
into the in vivo function of HIT proteins. RESULTS: The three-dimensional
crystal structures of free and nucleoside complexed FHIT have been determined
from multiwavelength anomalous diffraction (MAD) data, and they represent some
of the first successful structures to be measured with undulator radiation at
the Advanced Photon Source. The structures of FHIT reveal that this protein
exists as an intimate homodimer, which is based on a core structure observed
previously in another human HIT homolog, protein kinase C interacting protein
(PKCI), but has distinctive elaborations at both the N and C termini. Conserved
residues within the HIT family, which are involved in the interactions of the
proteins with nucleoside and phosphate groups, appear to be relevant for the
catalytic activity of this protein. CONCLUSIONS: The structure of FHIT, a
divergent HIT protein family member, in complex with a nucleotide analog
suggests a metal-independent catalytic mechanism for the HIT family of proteins.
A structural comparison of FHIT with PKCI and galactose-1-phosphate
uridylyltransferase (GaIT) reveals additional implications for the structural
and functional evolution of the ubiquitous HIT family of proteins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. Schematic stereo diagram of FHIT in
adenosine/sulfate product complexed form. Hydrogen-bonding
interactions thought to be important in catalysis and ligand
recognition are depicted by dotted lines. To simplify the
depiction, only a subset of residues have been shown
superimposed on the Ca backbone spline. Although most residues
involved in interactions between protein and adenine base are
not conserved between FHIT and PKCI (not shown here), both
proteins utilize a deep hydrophobic cleft and water-mediated
contacts to the backbone of N-terminal residues in binding the
adenosine base.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1997,
5,
763-774)
copyright 1997.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Martin,
M.V.St-Pierre,
and
J.F.Dufour
(2011).
Hit proteins, mitochondria and cancer.
|
| |
Biochim Biophys Acta,
1807,
626-632.
|
 |
|
|
|
|
 |
P.Tumbale,
C.D.Appel,
R.Kraehenbuehl,
P.D.Robertson,
J.S.Williams,
J.Krahn,
I.Ahel,
and
R.S.Williams
(2011).
Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease.
|
| |
Nat Struct Mol Biol,
18,
1189-1195.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.I.Hassan,
A.Naiyer,
and
F.Ahmad
(2010).
Fragile histidine triad protein: structure, function, and its association with tumorogenesis.
|
| |
J Cancer Res Clin Oncol,
136,
333-350.
|
 |
|
|
|
|
 |
M.K.Neyaz,
S.Hussain,
M.I.Hassan,
B.C.Das,
S.A.Husain,
and
M.Bharadwaj
(2010).
Novel missense mutation in FHIT gene: interpreting the effect in HPV-mediated cervical cancer in Indian women.
|
| |
Mol Cell Biochem,
335,
53-58.
|
 |
|
|
|
|
 |
U.Rass,
I.Ahel,
and
S.C.West
(2008).
Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin.
|
| |
J Biol Chem,
283,
33994-34001.
|
 |
|
|
|
|
 |
G.Rosenbaum,
R.W.Alkire,
G.Evans,
F.J.Rotella,
K.Lazarski,
R.G.Zhang,
S.L.Ginell,
N.Duke,
I.Naday,
J.Lazarz,
M.J.Molitsky,
L.Keefe,
J.Gonczy,
L.Rock,
R.Sanishvili,
M.A.Walsh,
E.Westbrook,
and
A.Joachimiak
(2006).
The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results.
|
| |
J Synchrotron Radiat,
13,
30-45.
|
 |
|
|
|
|
 |
W.T.Lo,
K.H.Chin,
H.L.Shr,
F.P.Gao,
P.C.Lyu,
A.H.Wang,
and
S.H.Chou
(2006).
Crystallization and preliminary X-ray analysis of XC1015, a histidine triad-like protein from Xanthomonas campestris.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1263-1265.
|
 |
|
|
|
|
 |
D.A.Kwasnicka,
A.Krakowiak,
C.Thacker,
C.Brenner,
and
S.R.Vincent
(2003).
Coordinate expression of NADPH-dependent flavin reductase, Fre-1, and Hint-related 7meGMP-directed hydrolase, DCS-1.
|
| |
J Biol Chem,
278,
39051-39058.
|
 |
|
|
|
|
 |
T.Su,
M.Suzui,
L.Wang,
C.S.Lin,
W.Q.Xing,
and
I.B.Weinstein
(2003).
Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis.
|
| |
Proc Natl Acad Sci U S A,
100,
7824-7829.
|
 |
|
|
|
|
 |
P.Bieganowski,
P.N.Garrison,
S.C.Hodawadekar,
G.Faye,
L.D.Barnes,
and
C.Brenner
(2002).
Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3.
|
| |
J Biol Chem,
277,
10852-10860.
|
 |
|
|
|
|
 |
S.Ingvarsson
(2001).
FHIT alterations in breast cancer.
|
| |
Semin Cancer Biol,
11,
361-366.
|
 |
|
|
|
|
 |
A.Draganescu,
S.C.Hodawadekar,
K.R.Gee,
and
C.Brenner
(2000).
Fhit-nucleotide specificity probed with novel fluorescent and fluorogenic substrates.
|
| |
J Biol Chem,
275,
4555-4560.
|
 |
|
|
|
|
 |
A.Guranowski
(2000).
Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates.
|
| |
Pharmacol Ther,
87,
117-139.
|
 |
|
|
|
|
 |
H.C.Pace,
S.C.Hodawadekar,
A.Draganescu,
J.Huang,
P.Bieganowski,
Y.Pekarsky,
C.M.Croce,
and
C.Brenner
(2000).
Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
|
| |
Curr Biol,
10,
907-917.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Shapiro,
and
T.Harris
(2000).
Finding function through structural genomics.
|
| |
Curr Opin Biotechnol,
11,
31-35.
|
 |
|
|
|
|
 |
A.Abend,
P.N.Garrison,
L.D.Barnes,
and
P.A.Frey
(1999).
Stereochemical retention of the configuration in the action of Fhit on phosphorus-chiral substrates.
|
| |
Biochemistry,
38,
3668-3676.
|
 |
|
|
|
|
 |
C.Brenner,
P.Bieganowski,
H.C.Pace,
and
K.Huebner
(1999).
The histidine triad superfamily of nucleotide-binding proteins.
|
| |
J Cell Physiol,
181,
179-187.
|
 |
|
|
|
|
 |
J.Elnatan,
D.Murphy,
H.S.Goh,
and
D.R.Smith
(1999).
HIT family genes: FHIT but not PKCI-1/HINT produces altered transcripts in colorectal cancer.
|
| |
Br J Cancer,
81,
874-880.
|
 |
|
|
|
|
 |
C.M.Ogata
(1998).
MAD phasing grows up.
|
| |
Nat Struct Biol,
5,
638-640.
|
 |
|
|
|
|
 |
H.C.Pace,
P.N.Garrison,
A.K.Robinson,
L.D.Barnes,
A.Draganescu,
A.Rösler,
G.M.Blackburn,
Z.Siprashvili,
C.M.Croce,
K.Huebner,
and
C.Brenner
(1998).
Genetic, biochemical, and crystallographic characterization of Fhit-substrate complexes as the active signaling form of Fhit.
|
| |
Proc Natl Acad Sci U S A,
95,
5484-5489.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Chen,
A.Brevet,
S.Blanquet,
and
P.Plateau
(1998).
Control of 5',5'-dinucleoside triphosphate catabolism by APH1, a Saccharomyces cerevisiae analog of human FHIT.
|
| |
J Bacteriol,
180,
2345-2349.
|
 |
|
|
|
|
 |
K.Huebner,
P.N.Garrison,
L.D.Barnes,
and
C.M.Croce
(1998).
The role of the FHIT/FRA3B locus in cancer.
|
| |
Annu Rev Genet,
32,
7.
|
 |
|
|
|
|
 |
C.Brenner,
H.C.Pace,
P.N.Garrison,
A.K.Robinson,
A.Rosler,
X.H.Liu,
G.M.Blackburn,
C.M.Croce,
K.Huebner,
and
L.D.Barnes
(1997).
Purification and crystallization of complexes modeling the active state of the fragile histidine triad protein.
|
| |
Protein Eng,
10,
1461-1463.
|
 |
|
|
|
|
 |
C.D.Lima,
M.G.Klein,
and
W.A.Hendrickson
(1997).
Structure-based analysis of catalysis and substrate definition in the HIT protein family.
|
| |
Science,
278,
286-290.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

