 |
PDBsum entry 2ffz

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.3
- phospholipase C.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phosphocholine + H2O = phosphocholine + a 1,2- diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
 1,2-diacyl-sn-glycero-3-phosphocholine
1,2-diacyl-sn-glycero-3-phosphocholine
|
+
|
H2O
|
=
|
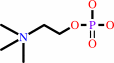 phosphocholine
phosphocholine
|
+
|
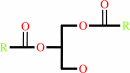 1,2- diacyl-sn-glycerol
1,2- diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Arch Biochem Biophys
460:41-47
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies examining the substrate specificity profiles of PC-PLC(Bc) protein variants.
|
|
A.P.Benfield,
N.M.Goodey,
L.T.Phillips,
S.F.Martin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The phosphatidylcholine preferring phospholipase C from Bacillus cereus
(PC-PLC(Bc)) catalyzes the hydrolysis of phospholipids in the following order of
preference: phosphatidylcholine (PC)>phosphatidylethanolamine
(PE)>phosphatidylserine (PS). In previous work, mutagenic, kinetic, and
crystallographic experiments suggested that varying the amino acids at the 4th,
56th, and 66th positions had a significant influence upon the substrate
specificity profile of PC-PLC(Bc). Here, we report the crystal structures of the
native form of several PC-PLC(Bc) variants that exhibited altered substrate
specificities for PC, PE, and PS at maximum resolutions of 1.90-2.05 Angstrom.
Comparing the structures of these variants to the structure of the wild-type
enzyme reveals only minor differences with respect to the number and location of
active site water molecules and the side chain conformations of residues at the
4th and 56th positions. These results suggest that subtle changes in steric and
electronic properties in the substrate binding site of PC-PLC(Bc) are
responsible for the significant changes in substrate selectivity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Karatsa-Dodgson,
M.E.Wörmann,
and
A.Gründling
(2010).
In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids.
|
| |
J Bacteriol,
192,
5341-5349.
|
 |
|
|
|
|
 |
P.Shindiapina,
and
C.Barlowe
(2010).
Requirements for transitional endoplasmic reticulum site structure and function in Saccharomyces cerevisiae.
|
| |
Mol Biol Cell,
21,
1530-1545.
|
 |
|
|
|
|
 |
S.G.Vachieri,
G.C.Clark,
A.Alape-Girón,
M.Flores-Díaz,
N.Justin,
C.E.Naylor,
R.W.Titball,
and
A.K.Basak
(2010).
Comparison of a nontoxic variant of Clostridium perfringens α-toxin with the toxic wild-type strain.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
1067-1074.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

