 |
PDBsum entry 2du7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.27
- O-phosphoserine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Cys) + O-phospho-L-serine + ATP = O-phospho-L-seryl-tRNA(Cys) + AMP + diphosphate
|
 |
 |
 |
 |
 |
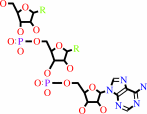 tRNA(Cys)
tRNA(Cys)
|
+
|
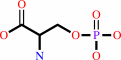 O-phospho-L-serine
O-phospho-L-serine
|
+
|
 ATP
ATP
|
=
|
O-phospho-L-seryl-tRNA(Cys)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
14:272-279
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea.
|
|
R.Fukunaga,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cysteine is ligated to tRNA(Cys) by cysteinyl-tRNA synthetase in most organisms.
However, in methanogenic archaea lacking cysteinyl-tRNA synthetase,
O-phosphoserine is ligated to tRNA(Cys) by O-phosphoseryl-tRNA synthetase
(SepRS), and the phosphoseryl-tRNA(Cys) is converted to cysteinyl-tRNA(Cys). In
this study, we determined the crystal structure of the SepRS tetramer in complex
with tRNA(Cys) and O-phosphoserine at 2.6-A resolution. The catalytic domain of
SepRS recognizes the negatively charged side chain of O-phosphoserine at a
noncanonical site, using the dipole moment of a conserved alpha-helix. The
unique C-terminal domain specifically recognizes the anticodon GCA of tRNA(Cys).
On the basis of the structure, we engineered SepRS to recognize tRNA(Cys)
mutants with the anticodons UCA and CUA and clarified the anticodon recognition
mechanism by crystallography. The mutant SepRS-tRNA pairs may be useful for
translational incorporation of O-phosphoserine into proteins in response to the
stop codons UGA and UAG.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
(a) Superposition of the catalytic domains of SepRS (pink)
and PheRS (light blue). Cyan ball-and-stick model,
O-phosphoserine; gray ball-and-stick, phenylalanine; dark blue,
magenta and violet tubes, motifs 1, 2 and 3, respectively, which
are characteristic of class II aaRSs; dark green, central helix
and the preceding loop. (b) The catalytic site in the A.
fulgidus SepRS–tRNA^Cys–O-phosphoserine complex. Yellow,
O-phosphoserine carbons; cyan, O-phosphoserine phosphorus atoms.
SepRS is colored as in a. (c) The catalytic site in the T.
thermophilus PheRS–phenylalanine complex, colored as in b. (d)
Schematic representation of the unique recognition mechanism for
O-phosphoserine by SepRS.
|
 |
Figure 3.
(a) Structure of the anticodon-binding domain (stereo view).
Green, helices; blue,  -strands;
yellow, tRNA; red, anticodon nucleotides; ball-and-stick models,
Glu418 and Glu420. (b) The recognition of the tRNA^Cys anticodon
loop in the SepRS–tRNA^Cys–O-phosphoserine complex (stereo
view). Pink and green mesh, |F[o] - F[c]| simulated-annealing
omit electron density maps (3.0 -strands;
yellow, tRNA; red, anticodon nucleotides; ball-and-stick models,
Glu418 and Glu420. (b) The recognition of the tRNA^Cys anticodon
loop in the SepRS–tRNA^Cys–O-phosphoserine complex (stereo
view). Pink and green mesh, |F[o] - F[c]| simulated-annealing
omit electron density maps (3.0  )
for the tRNA anticodon loop nucleotides and the SepRS
recognition residues, respectively. (c) Recognition of A36, G37
and A38. (d) Recognition of G34 and C35. Pink, the two residues
(Glu418 and Glu420) that were mutated for engineering. )
for the tRNA anticodon loop nucleotides and the SepRS
recognition residues, respectively. (c) Recognition of A36, G37
and A38. (d) Recognition of G34 and C35. Pink, the two residues
(Glu418 and Glu420) that were mutated for engineering.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2007,
14,
272-279)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Finarov,
N.Moor,
N.Kessler,
L.Klipcan,
and
M.G.Safro
(2010).
Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
|
| |
Structure,
18,
343-353.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
C.M.Zhang,
C.Liu,
S.Slater,
and
Y.M.Hou
(2008).
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys).
|
| |
Nat Struct Mol Biol,
15,
507-514.
|
 |
|
|
|
|
 |
J.Yuan,
K.Sheppard,
and
D.Söll
(2008).
Amino acid modifications on tRNA.
|
| |
Acta Biochim Biophys Sin (Shanghai),
40,
539-553.
|
 |
|
|
|
|
 |
K.Sheppard,
J.Yuan,
M.J.Hohn,
B.Jester,
K.M.Devine,
and
D.Söll
(2008).
From one amino acid to another: tRNA-dependent amino acid biosynthesis.
|
| |
Nucleic Acids Res,
36,
1813-1825.
|
 |
|
|
|
|
 |
L.Klipcan,
I.Levin,
N.Kessler,
N.Moor,
I.Finarov,
and
M.Safro
(2008).
The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase.
|
| |
Structure,
16,
1095-1104.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Goto-Ito,
T.Ito,
R.Ishii,
Y.Muto,
Y.Bessho,
and
S.Yokoyama
(2008).
Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5.
|
| |
Proteins,
72,
1274-1289.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.I.Hauenstein,
and
J.J.Perona
(2008).
Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei.
|
| |
J Biol Chem,
283,
22007-22017.
|
 |
|
|
|
|
 |
S.I.Hauenstein,
Y.M.Hou,
and
J.J.Perona
(2008).
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.
|
| |
J Biol Chem,
283,
21997-22006.
|
 |
|
|
|
|
 |
J.M.Kavran,
S.Gundllapalli,
P.O'Donoghue,
M.Englert,
D.Söll,
and
T.A.Steitz
(2007).
Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
|
| |
Proc Natl Acad Sci U S A,
104,
11268-11273.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

