
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.2.3.2.2
- gamma-glutamyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-terminal (5-L-glutamyl)-[peptide] + an alpha-amino acid = 5-L- glutamyl amino acid + an N-terminal L-alpha-aminoacyl-[peptide]
|
 |
 |
 |
 |
 |
N-terminal (5-L-glutamyl)-[peptide]
|
+
|
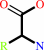 alpha-amino acid
alpha-amino acid
|
=
|
 5-L- glutamyl amino acid
5-L- glutamyl amino acid
|
+
|
N-terminal L-alpha-aminoacyl-[peptide]
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.3.4.19.13
- glutathione gamma-glutamate hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glutathione + H2O = L-cysteinylglycine + L-glutamate
|
|
2.
|
an S-substituted glutathione + H2O = an S-substituted L-cysteinylglycine + L-glutamate
|
|
 |
 |
 |
 |
 |
 glutathione
glutathione
|
+
|
H2O
|
=
|
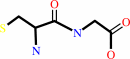 L-cysteinylglycine
L-cysteinylglycine
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
S-substituted glutathione
|
+
|
H2O
|
=
|
S-substituted L-cysteinylglycine
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:6471-6476
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
|
|
T.Okada,
H.Suzuki,
K.Wada,
H.Kumagai,
K.Fukuyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Gamma-glutamyltranspeptidase (GGT) is a heterodimic enzyme that is generated
from the precursor protein through posttranslational processing and catalyzes
the hydrolysis of gamma-glutamyl bonds in gamma-glutamyl compounds such as
glutathione and/or the transfer of the gamma-glutamyl group to other amino acids
and peptides. We have determined the crystal structure of GGT from Escherichia
coli K-12 at 1.95 A resolution. GGT has a stacked alphabetabetaalpha fold
comprising the large and small subunits, similar to the folds seen in members of
the N-terminal nucleophile hydrolase superfamily. The active site Thr-391, the
N-terminal residue of the small subunit, is located in the groove, from which
the pocket for gamma-glutamyl moiety binding follows. We have further determined
the structure of the gamma-glutamyl-enzyme intermediate trapped by flash cooling
the GGT crystal soaked in glutathione solution and the structure of GGT in
complex with l-glutamate. These structures revealed how the gamma-glutamyl
moiety and l-glutamate are recognized by the enzyme. A water molecule was seen
on the carbonyl carbon of the gamma-glutamyl-Thr-391 Ogamma bond in the
intermediate that is to be hydrolyzed. Notably the residues essential for GGT
activity (Arg-114, Asp-433, Ser-462, and Ser-463 in E. coli GGT) shown by
site-directed mutagenesis of human GGT are all involved in the binding of the
gamma-glutamyl moiety. The structure of E. coli GGT presented here, together
with sequence alignment of GGTs, may be applicable to interpret the biochemical
and genetic data of other GGTs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Structure of E. coli GGT. (a) Ribbon drawing of the
GGT heterodimer. The L subunit is colored blue, and the S
subunit is colored green. (b) Ribbon drawing of the L subunit.
(c) Ribbon drawing of the S subunit.  -Helices and -Helices and  -strands
are labeled. In each of the L and S subunits, the N terminus is
blue and the C terminus is red, with intermediate colors
following the distance in the sequence from the N terminus. The
N-terminal residue of the S subunit (Thr-391) is shown with a
stick model. (d) A topology diagram of E. coli GGT. Circle,
triangle, and square indicate -strands
are labeled. In each of the L and S subunits, the N terminus is
blue and the C terminus is red, with intermediate colors
following the distance in the sequence from the N terminus. The
N-terminal residue of the S subunit (Thr-391) is shown with a
stick model. (d) A topology diagram of E. coli GGT. Circle,
triangle, and square indicate  -helix, -helix,  -strand,
and insertion not conserved among Ntn-hydrolases, respectively.
The secondary structures were defined with DSSP (19). The
figures were prepared with PYMOL (20) and TOPS (21). -strand,
and insertion not conserved among Ntn-hydrolases, respectively.
The secondary structures were defined with DSSP (19). The
figures were prepared with PYMOL (20) and TOPS (21).
|
 |
Figure 2.
Fig. 2. The structure of the substrate binding pocket of E.
coli GGT. (a) Surface drawing of substrate binding pocket. The
stick model of the  -glutamyl moiety,
nucleophile (Thr-391), and residues forming the wall (Asn-411
and Tyr-444) are shown in blue, green, and yellow, respectively.
Green dots represent the groove in which the peptide of the
precursor protein is assumed to be present. The hydrogen bond
between Asn-411 O -glutamyl moiety,
nucleophile (Thr-391), and residues forming the wall (Asn-411
and Tyr-444) are shown in blue, green, and yellow, respectively.
Green dots represent the groove in which the peptide of the
precursor protein is assumed to be present. The hydrogen bond
between Asn-411 O  and Tyr-444 O and Tyr-444 O  is shown
as a dashed line. The ribbon model shown in yellow represents
residues Pro-438–Gly-449, which are absent in B. subtilis GGT.
(b) The (F[o] – F[c]) omit map contoured at the 3 is shown
as a dashed line. The ribbon model shown in yellow represents
residues Pro-438–Gly-449, which are absent in B. subtilis GGT.
(b) The (F[o] – F[c]) omit map contoured at the 3  level
for GGT- level
for GGT-  G. The omit map was
generated by omitting the G. The omit map was
generated by omitting the  -glutamyl moiety,
Thr-391, and a water molecule (labeled W2) from the model.
Ball-and-stick models of -glutamyl moiety,
Thr-391, and a water molecule (labeled W2) from the model.
Ball-and-stick models of  -glutamyl–enzyme
complex are overlaid on the map. The residues involved in
substrate binding and enzyme reaction are shown in the model.
For the clarity, the side chains of Gln-89, Leu-410, and Thr-412
are omitted from the model. Water molecules involved in
substrate binding and the catalytic reaction are labeled (W1 -glutamyl–enzyme
complex are overlaid on the map. The residues involved in
substrate binding and enzyme reaction are shown in the model.
For the clarity, the side chains of Gln-89, Leu-410, and Thr-412
are omitted from the model. Water molecules involved in
substrate binding and the catalytic reaction are labeled (W1
 W3).
The hydrogen bonds are shown as dashed lines. (c) The (F[o] –
F[c]) omit map for GGT-Glu prepared as for GGT- W3).
The hydrogen bonds are shown as dashed lines. (c) The (F[o] –
F[c]) omit map for GGT-Glu prepared as for GGT-  G. The
view direction is rotated by 40° around the vertical axis
relative to b. The figures were prepared with PYMOL (20). G. The
view direction is rotated by 40° around the vertical axis
relative to b. The figures were prepared with PYMOL (20).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Castellano,
A.Di Salle,
A.Merlino,
M.Rossi,
and
F.La Cara
(2011).
Gene cloning and protein expression of γ-glutamyltranspeptidases from Thermus thermophilus and Deinococcus radiodurans: comparison of molecular and structural properties with mesophilic counterparts.
|
| |
Extremophiles,
15,
259-270.
|
 |
|
|
|
|
 |
F.H.Hausheer,
D.Shanmugarajah,
B.D.Leverett,
X.Chen,
Q.Huang,
H.Kochat,
P.N.Petluru,
and
A.R.Parker
(2010).
Mechanistic study of BNP7787-mediated cisplatin nephroprotection: modulation of gamma-glutamyl transpeptidase.
|
| |
Cancer Chemother Pharmacol,
65,
941-951.
|
 |
|
|
|
|
 |
F.Zhang,
Q.Z.Zheng,
Q.C.Jiao,
J.Z.Liu,
and
G.H.Zhao
(2010).
Enzymatic synthesis of theanine from glutamic acid γ-methyl ester and ethylamine by immobilized Escherichia coli cells with γ-glutamyltranspeptidase activity.
|
| |
Amino Acids,
39,
1177-1182.
|
 |
|
|
|
|
 |
H.P.Chang,
W.C.Liang,
R.C.Lyu,
M.C.Chi,
T.F.Wang,
K.L.Su,
H.C.Hung,
and
L.L.Lin
(2010).
Effects of C-terminal truncation on autocatalytic processing of Bacillus licheniformis gamma-glutamyl transpeptidase.
|
| |
Biochemistry (Mosc),
75,
919-929.
|
 |
|
|
|
|
 |
H.Suzuki,
C.Yamada,
K.Kijima,
S.Ishihara,
K.Wada,
K.Fukuyama,
and
H.Kumagai
(2010).
Enhancement of glutaryl-7-aminocephalosporanic acid acylase activity of gamma-glutamyltranspeptidase of Bacillus subtilis.
|
| |
Biotechnol J,
5,
829-837.
|
 |
|
|
|
|
 |
K.Wada,
M.Irie,
H.Suzuki,
and
K.Fukuyama
(2010).
Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket.
|
| |
FEBS J,
277,
1000-1009.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Bokhove,
P.N.Jimenez,
W.J.Quax,
and
B.W.Dijkstra
(2010).
The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket.
|
| |
Proc Natl Acad Sci U S A,
107,
686-691.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Zhiryakova,
I.Ivanov,
S.Ilieva,
M.Guncheva,
B.Galunsky,
and
N.Stambolieva
(2009).
Do N-terminal nucleophile hydrolases indeed have a single amino acid catalytic center?
|
| |
FEBS J,
276,
2589-2598.
|
 |
|
|
|
|
 |
J.B.King,
M.B.West,
P.F.Cook,
and
M.H.Hanigan
(2009).
A novel, species-specific class of uncompetitive inhibitors of gamma-glutamyl transpeptidase.
|
| |
J Biol Chem,
284,
9059-9065.
|
 |
|
|
|
|
 |
K.Williams,
S.Cullati,
A.Sand,
E.I.Biterova,
and
J.J.Barycki
(2009).
Crystal structure of acivicin-inhibited gamma-glutamyltranspeptidase reveals critical roles for its C-terminus in autoprocessing and catalysis.
|
| |
Biochemistry,
48,
2459-2467.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.C.Lyu,
H.Y.Hu,
L.Y.Kuo,
H.F.Lo,
P.L.Ong,
H.P.Chang,
and
L.L.Lin
(2009).
Role of the conserved Thr399 and Thr417 residues of Bacillus licheniformis gamma-Glutamyltranspeptidase as evaluated by mutational analysis.
|
| |
Curr Microbiol,
59,
101-106.
|
 |
|
|
|
|
 |
R.Wu,
S.Richter,
R.G.Zhang,
V.J.Anderson,
D.Missiakas,
and
A.Joachimiak
(2009).
Crystal structure of Bacillus anthracis transpeptidase enzyme CapD.
|
| |
J Biol Chem,
284,
24406-24414.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Richter,
V.J.Anderson,
G.Garufi,
L.Lu,
J.M.Budzik,
A.Joachimiak,
C.He,
O.Schneewind,
and
D.Missiakas
(2009).
Capsule anchoring in Bacillus anthracis occurs by a transpeptidation reaction that is inhibited by capsidin.
|
| |
Mol Microbiol,
71,
404-420.
|
 |
|
|
|
|
 |
C.Yamada,
K.Kijima,
S.Ishihara,
C.Miwa,
K.Wada,
T.Okada,
K.Fukuyama,
H.Kumagai,
and
H.Suzuki
(2008).
Improvement of the glutaryl-7-aminocephalosporanic acid acylase activity of a bacterial gamma-glutamyltranspeptidase.
|
| |
Appl Environ Microbiol,
74,
3400-3409.
|
 |
|
|
|
|
 |
N.Heisterkamp,
J.Groffen,
D.Warburton,
and
T.P.Sneddon
(2008).
The human gamma-glutamyltransferase gene family.
|
| |
Hum Genet,
123,
321-332.
|
 |
|
|
|
|
 |
R.Zarnowski,
K.G.Cooper,
L.S.Brunold,
J.Calaycay,
and
J.P.Woods
(2008).
Histoplasma capsulatum secreted gamma-glutamyltransferase reduces iron by generating an efficient ferric reductant.
|
| |
Mol Microbiol,
70,
352-368.
|
 |
|
|
|
|
 |
H.Suzuki,
C.Yamada,
and
K.Kato
(2007).
Gamma-glutamyl compounds and their enzymatic production using bacterial gamma-glutamyltranspeptidase.
|
| |
Amino Acids,
32,
333-340.
|
 |
|
|
|
|
 |
H.Xie,
S.Vucetic,
L.M.Iakoucheva,
C.J.Oldfield,
A.K.Dunker,
Z.Obradovic,
and
V.N.Uversky
(2007).
Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins.
|
| |
J Proteome Res,
6,
1917-1932.
|
 |
|
|
|
|
 |
K.Shibayama,
J.Wachino,
Y.Arakawa,
M.Saidijam,
N.G.Rutherford,
and
P.J.Henderson
(2007).
Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism.
|
| |
Mol Microbiol,
64,
396-406.
|
 |
|
|
|
|
 |
M.Morin,
C.Rivard,
and
J.W.Keillor
(2006).
gamma-Glutamyl transpeptidase acylation with peptidic substrates: free energy relationships measured by an HPLC kinetic assay.
|
| |
Org Biomol Chem,
4,
3790-3801.
|
 |
|
|
|
|
 |
Y.F.Yao,
Y.M.Weng,
H.Y.Hu,
K.L.Ku,
and
L.L.Lin
(2006).
Expression optimization and biochemical characterization of a recombinant gamma-glutamyltranspeptidase from Escherichia coli novablue.
|
| |
Protein J,
25,
431-441.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

