 |
PDBsum entry 2cl5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Catechol-o-methyltransferase in complex with an inhibitor
|
|
Structure:
|
 |
Catechol o-methyltransferase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Rattus norvegicus. Rat. Organism_taxid: 10116. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.151
|
R-free:
|
0.178
|
|
|
Authors:
|
 |
P.N.Palma,M.L.Rodrigues,M.Archer,M.J.Bonifacio,A.I.Loureiro, D.A.Learmonth,M.A.Carrondo,P.Soares-Da-Silva
|
|
Key ref:
|
 |
P.N.Palma
et al.
(2006).
Comparative study of ortho- and meta-nitrated inhibitors of catechol-O-methyltransferase: interactions with the active site and regioselectivity of O-methylation.
Mol Pharmacol,
70,
143-153.
PubMed id:
|
 |
|
Date:
|
 |
|
26-Apr-06
|
Release date:
|
28-Jun-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P22734
(COMT_RAT) -
Catechol O-methyltransferase from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
264 a.a.
215 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.6
- catechol O-methyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a catechol + S-adenosyl-L-methionine = a guaiacol + S-adenosyl-L- homocysteine + H+
|
 |
 |
 |
 |
 |
catechol
Bound ligand (Het Group name = )
matches with 42.11% similarity
|
+
|
S-adenosyl-L-methionine
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
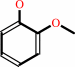 guaiacol
guaiacol
|
+
|
 S-adenosyl-L- homocysteine
S-adenosyl-L- homocysteine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Mol Pharmacol
70:143-153
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Comparative study of ortho- and meta-nitrated inhibitors of catechol-O-methyltransferase: interactions with the active site and regioselectivity of O-methylation.
|
|
P.N.Palma,
M.L.Rodrigues,
M.Archer,
M.J.Bonifácio,
A.I.Loureiro,
D.A.Learmonth,
M.A.Carrondo,
P.Soares-da-Silva.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In this work, we present a comparative case study of "ortho-" and
"meta-nitrated" catecholic inhibitors of catechol-O-methyltransferase (COMT),
with regard to their interaction with the catalytic site of the enzyme and the
in vitro regioselective formation of their mono-O-methyl ether metabolites. In
particular, the effects of altering the attachment position of the inhibitors'
side-chain substituent, within the classic nitrocatechol pharmacophore, were
investigated. For this purpose, we compared two simple regioisomeric
nitrocatechol-type inhibitors of COMT, BIA 3-228 and BIA 8-176, which contain
the benzoyl substituent attached at the meta and ortho positions, respectively,
relative to the nitro group. The two compounds were slowly O-methylated by COMT
in vitro, but the particular substitution pattern of each compound was shown to
have a profound impact on the regioselectivity of their O-methylation. To
provide a plausible interpretation of these results, a comprehensive analysis of
the protein-inhibitor interactions and of the relative chemical susceptibility
to O-methylation of the catechol hydroxyl groups was performed by means of
docking simulations and ab initio molecular orbital calculations. The major
structural and chemical factors that determine the enzyme regioselectivity of
O-methylation were identified, and the X-ray structure of the complex of COMT
with S-adenosyl-l-methionine and BIA 8-176 is herein disclosed. This is the
first reported structure of the soluble form of COMT complexed with a
nitrocatecholic inhibitor having a bulky substituent group in adjacent position
(ortho) to the nitro group. Structural and dynamic aspects of this complex are
analyzed and discussed, in the context of the present study.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Gómez García,
C.E.Stevenson,
I.Usón,
C.L.Freel Meyers,
C.T.Walsh,
and
D.M.Lawson
(2010).
The crystal structure of the novobiocin biosynthetic enzyme NovP: the first representative structure for the TylF O-methyltransferase superfamily.
|
| |
J Mol Biol,
395,
390-407.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Singh,
J.G.McCoy,
C.Zhang,
C.A.Bingman,
G.N.Phillips,
and
J.S.Thorson
(2008).
Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
|
| |
J Biol Chem,
283,
22628-22636.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Bonifácio,
P.N.Palma,
L.Almeida,
and
P.Soares-da-Silva
(2007).
Catechol-O-methyltransferase and its inhibitors in Parkinson's disease.
|
| |
CNS Drug Rev,
13,
352-379.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

