 |
PDBsum entry 2cis

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.3.15
- glucose-6-phosphate 1-epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose 6-phosphate = beta-D-glucose 6-phosphate
|
 |
 |
 |
 |
 |
alpha-D-glucose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
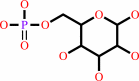 beta-D-glucose 6-phosphate
beta-D-glucose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:30175-30185
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
|
|
M.Graille,
J.P.Baltaze,
N.Leulliot,
D.Liger,
S.Quevillon-Cheruel,
H.van Tilbeurgh.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Despite the generation of a large amount of sequence information over the last
decade, more than 40% of well characterized enzymatic functions still lack
associated protein sequences. Assigning protein sequences to documented
biochemical functions is an interesting challenge. We illustrate here that
structural genomics may be a reasonable approach in addressing these questions.
We present the crystal structure of the Saccharomyces cerevisiae YMR099cp, a
protein of unknown function. YMR099cp adopts the same fold as galactose
mutarotase and shares the same catalytic machinery necessary for the
interconversion of the alpha and beta anomers of galactose. The structure
revealed the presence in the active site of a sulfate ion attached by an
arginine clamp made by the side chain from two strictly conserved arginine
residues. This sulfate is ideally positioned to mimic the phosphate group of
hexose 6-phosphate. We have subsequently successfully demonstrated that YMR099cp
is a hexose-6-phosphate mutarotase with broad substrate specificity. We solved
high resolution structures of some substrate enzyme complexes, further
confirming our functional hypothesis. The metabolic role of a hexose-6-phosphate
mutarotase is discussed. This work illustrates that structural information has
been crucial to assign YMR099cp to the orphan EC activity: hexose-phosphate
mutarotase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Schematic representations of phosphosugars used in this study.
|
 |
Figure 6.
Complexes of YMR099cp bound to hexose 6-phosphate sugars.
Stereoview representation of Glc6P (A) and Tag6P (B) bound into
the YMR099cp active site. The 2F[o] - F[c] electron density maps
contoured at 1σ are shown in blue around the ligands. Hydrogen
bonds made by the ligands with YMR099cp as well as Ba^2+ Tag6P
oxygen coordination are depicted by red dashed lines. Water
molecules are shown by red spheres.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
30175-30185)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Mukherjee,
and
Y.Zhang
(2009).
MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming.
|
| |
Nucleic Acids Res,
37,
e83.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

