
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Ribonucleotide reductase class 1b holocomplex r1e,r2f from salmonella typhimurium
|
|
Structure:
|
 |
Ribonucleoside-diphosphate reductase 2 alpha subunit. Chain: e, f. Synonym: ribonucleotide reductase 2, r1e protein. Engineered: yes. Ribonucleoside-diphosphate reductase 2 beta subunit. Chain: i, j. Synonym: ribonucleotide reductase 2, r2f protein. Engineered: yes
|
|
Source:
|
 |
Salmonella typhimurium. Organism_taxid: 602. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
3.99Å
|
R-factor:
|
0.265
|
R-free:
|
0.310
|
|
|
Authors:
|
 |
M.Uppsten,M.Farnegardh,V.Domkin,U.Uhlin
|
Key ref:

|
 |
M.Uppsten
et al.
(2006).
The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action.
J Mol Biol,
359,
365-377.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Apr-05
|
Release date:
|
17-May-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains E, F, I, J:
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
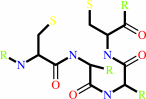
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
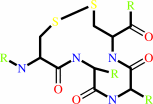 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
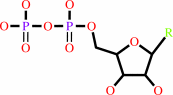
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
359:365-377
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action.
|
|
M.Uppsten,
M.Färnegårdh,
V.Domkin,
U.Uhlin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ribonucleotide reductase is an indispensable enzyme for all cells, since it
catalyses the biosynthesis of the precursors necessary for both building and
repairing DNA. The ribonucleotide reductase class I enzymes, present in all
mammals as well as in many prokaryotes and DNA viruses, are composed mostly of
two homodimeric proteins, R1 and R2. The reaction involves long-range radical
transfer between the two proteins. Here, we present the first crystal structure
of a ribonucleotide reductase R1/R2 holocomplex. The biological relevance of
this complex is based on the binding of the R2 C terminus in the hydrophobic
cleft of R1, an interaction proven to be crucial for enzyme activity, and by the
fact that all conserved amino acid residues in R2 are facing the R1 active
sites. We suggest that the asymmetric R1/R2 complex observed in the 4A crystal
structure of Salmonella typhimurium ribonucleotide reductase represents an
intermediate stage in the reaction cycle, and at the moment of reaction the
homodimers transiently form a tight symmetric complex.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. The R2F protein. (a) The R2F dimer structure with the
conserved side-chains and iron atoms highlighted. One monomer
is in dark blue and the other is in blue-grey. Conserved
side-chains are shown as yellow sticks. The iron atoms are shown
as orange spheres. (b) The R2F iron-centre, with the iron atoms
(orange) built into the electron density. The 2F[o]−F[c]
density map contoured at 1.0σ (blue) and the F[o]−F[c] omit
map contoured at 2.6σ (green). Figure 2. The R2F protein.
(a) The R2F dimer structure with the conserved side-chains and
iron atoms highlighted. One monomer is in dark blue and the
other is in blue-grey. Conserved side-chains are shown as yellow
sticks. The iron atoms are shown as orange spheres. (b) The R2F
iron-centre, with the iron atoms (orange) built into the
electron density. The 2F[o]−F[c] density map contoured at
1.0σ (blue) and the F[o]−F[c] omit map contoured at 2.6σ
(green).
|
 |
Figure 3.
Figure 3. The R1E protein. (a) The R1E dimer structure. The
N-terminal domains are coloured in blue-green, the small domains
in gold. The catalytic ten-stranded α/β barrel is coloured
brown in one monomer and beige in the other. One hydrophobic
cleft of R1E is marked (blue peptide) as well as the helices
forming the cleft. The radical forming C388 at the active sites
are marked as spheres coloured by atom. The dGTP effectors on
each side of the dimer interface are shown in sticks. Three
loops are marked. Flexible parts of the loops are indicated by
broken lines. (b)The effector dGTP at the specificity site of
R1E. In brown is the initial F[o]−F[c] electron density map
contoured at 3.0σ and in light grey the final 2F[o]−F[c]
density map contoured at 1.0σ. The loop shown in the picture,
loop 1, is stabilised by the binding of the effector. The
magnesium ion is colored magenta with the F[o]−F[c] omit map
for the Mg in green (contoured at 3.0σ). Figure 3. The R1E
protein. (a) The R1E dimer structure. The N-terminal domains are
coloured in blue-green, the small domains in gold. The catalytic
ten-stranded α/β barrel is coloured brown in one monomer and
beige in the other. One hydrophobic cleft of R1E is marked (blue
peptide) as well as the helices forming the cleft. The radical
forming C388 at the active sites are marked as spheres coloured
by atom. The dGTP effectors on each side of the dimer interface
are shown in sticks. Three loops are marked. Flexible parts of
the loops are indicated by broken lines. (b)The effector dGTP at
the specificity site of R1E. In brown is the initial F[o]−F[c]
electron density map contoured at 3.0σ and in light grey the
final 2F[o]−F[c] density map contoured at 1.0σ. The loop
shown in the picture, loop 1, is stabilised by the binding of
the effector. The magnesium ion is colored magenta with the
F[o]−F[c] omit map for the Mg in green (contoured at 3.0σ).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
359,
365-377)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.T.Logan
(2011).
Closing the circle on ribonucleotide reductases.
|
| |
Nat Struct Mol Biol,
18,
251-253.
|
 |
|
|
|
|
 |
J.W.Fairman,
S.R.Wijerathna,
M.F.Ahmad,
H.Xu,
R.Nakano,
S.Jha,
J.Prendergast,
R.M.Welin,
S.Flodin,
A.Roos,
P.Nordlund,
Z.Li,
T.Walz,
and
C.G.Dealwis
(2011).
Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
|
| |
Nat Struct Mol Biol,
18,
316-322.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Boal,
J.A.Cotruvo,
J.Stubbe,
and
A.C.Rosenzweig
(2010).
Structural basis for activation of class Ib ribonucleotide reductase.
|
| |
Science,
329,
1526-1530.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.B.Gammon,
B.Gowrishankar,
S.Duraffour,
G.Andrei,
C.Upton,
and
D.H.Evans
(2010).
Vaccinia virus-encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis.
|
| |
PLoS Pathog,
6,
e1000984.
|
 |
|
|
|
|
 |
A.R.Offenbacher,
I.R.Vassiliev,
M.R.Seyedsayamdost,
J.Stubbe,
and
B.A.Barry
(2009).
Redox-linked structural changes in ribonucleotide reductase.
|
| |
J Am Chem Soc,
131,
7496-7497.
|
 |
|
|
|
|
 |
J.Wang,
G.J.Lohman,
and
J.Stubbe
(2009).
Mechanism of inactivation of human ribonucleotide reductase with p53R2 by gemcitabine 5'-diphosphate.
|
| |
Biochemistry,
48,
11612-11621.
|
 |
|
|
|
|
 |
A.Q.Hassan,
and
J.Stubbe
(2008).
Mapping the subunit interface of ribonucleotide reductase (RNR) using photo cross-linking.
|
| |
Bioorg Med Chem Lett,
18,
5923-5925.
|
 |
|
|
|
|
 |
A.Q.Hassan,
Y.Wang,
L.Plate,
and
J.Stubbe
(2008).
Methodology to probe subunit interactions in ribonucleotide reductases.
|
| |
Biochemistry,
47,
13046-13055.
|
 |
|
|
|
|
 |
H.Xu,
J.W.Fairman,
S.R.Wijerathna,
N.R.Kreischer,
J.LaMacchia,
E.Helmbrecht,
B.S.Cooperman,
and
C.Dealwis
(2008).
The structural basis for peptidomimetic inhibition of eukaryotic ribonucleotide reductase: a conformationally flexible pharmacophore.
|
| |
J Med Chem,
51,
4653-4659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Nurbo,
A.K.Roos,
D.Muthas,
E.Wahlström,
D.J.Ericsson,
T.Lundstedt,
T.Unge,
and
A.Karlén
(2007).
Design, synthesis and evaluation of peptide inhibitors of Mycobacterium tuberculosis ribonucleotide reductase.
|
| |
J Pept Sci,
13,
822-832.
|
 |
|
|
|
|
 |
J.Wang,
G.J.Lohman,
and
J.Stubbe
(2007).
Enhanced subunit interactions with gemcitabine-5'-diphosphate inhibit ribonucleotide reductases.
|
| |
Proc Natl Acad Sci U S A,
104,
14324-14329.
|
 |
|
|
|
|
 |
A.J.Narváez,
N.Voevodskaya,
L.Thelander,
and
A.Gräslund
(2006).
The involvement of Arg265 of mouse ribonucleotide reductase R2 protein in proton transfer and catalysis.
|
| |
J Biol Chem,
281,
26022-26028.
|
 |
|
|
|
|
 |
M.Galander,
M.Uppsten,
U.Uhlin,
and
F.Lendzian
(2006).
Orientation of the tyrosyl radical in Salmonella typhimurium class Ib ribonucleotide reductase determined by high field EPR of R2F single crystals.
|
| |
J Biol Chem,
281,
31743-31752.
|
 |
|
|
|
|
 |
V.P.Denysenkov,
T.F.Prisner,
J.Stubbe,
and
M.Bennati
(2006).
High-field pulsed electron-electron double resonance spectroscopy to determine the orientation of the tyrosyl radicals in ribonucleotide reductase.
|
| |
Proc Natl Acad Sci U S A,
103,
13386-13390.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

