 |
PDBsum entry 2b5h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2b5h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.20
- cysteine dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteine + O2 = 3-sulfino-L-alanine + H+
|
 |
 |
 |
 |
 |
 L-cysteine
L-cysteine
|
+
|
 O2
O2
|
=
|
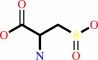 3-sulfino-L-alanine
3-sulfino-L-alanine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); NADH or NADPH
|
 |
 |
 |
 |
 |
Fe(2+)
|
 NADH
NADH
|
or
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:18723-18733
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of mammalian cysteine dioxygenase. A novel mononuclear iron center for cysteine thiol oxidation.
|
|
C.R.Simmons,
Q.Liu,
Q.Huang,
Q.Hao,
T.P.Begley,
P.A.Karplus,
M.H.Stipanuk.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cysteine dioxygenase is a mononuclear iron-dependent enzyme responsible for the
oxidation of cysteine with molecular oxygen to form cysteine sulfinate. This
reaction commits cysteine to either catabolism to sulfate and pyruvate or the
taurine biosynthetic pathway. Cysteine dioxygenase is a member of the cupin
superfamily of proteins. The crystal structure of recombinant rat cysteine
dioxygenase has been determined to 1.5-A resolution, and these results confirm
the canonical cupin beta-sandwich fold and the rare cysteinyltyrosine
intramolecular cross-link (between Cys(93) and Tyr(157)) seen in the recently
reported murine cysteine dioxygenase structure. In contrast to the catalytically
inactive mononuclear Ni(II) metallocenter present in the murine structure,
crystallization of a catalytically competent preparation of rat cysteine
dioxygenase revealed a novel tetrahedrally coordinated mononuclear iron center
involving three histidines (His(86), His(88), and His(140)) and a water
molecule. Attempts to acquire a structure with bound ligand using either
cocrystallization or soaking crystals with cysteine revealed the formation of a
mixed disulfide involving Cys(164) near the active site, which may explain
previously observed substrate inhibition. This work provides a framework for
understanding the molecular mechanisms involved in thiol dioxygenation and sets
the stage for exploration of the chemistry of both the novel mononuclear iron
center and the catalytic role of the cysteinyl-tyrosine linkage.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Electron density evidence for key features of the
CDO active site. 2F[o] - F[c] electron density is shown
contoured at 1.6  . Stick representations
of select protein residues, including the Cys-Tyr linkage, are
shown with iron (orange sphere) and active site waters (red
spheres). All structural figures within this report were
prepared using PyMOL (62). . Stick representations
of select protein residues, including the Cys-Tyr linkage, are
shown with iron (orange sphere) and active site waters (red
spheres). All structural figures within this report were
prepared using PyMOL (62).
|
 |
Figure 8.
FIGURE 8. Mechanistic proposals for CDO. Scheme A,
mechanistic proposal for the catalytic cycle of cysteine
oxidation. The letter B in this Scheme A indicates a putative
active site base. Scheme B, mechanistic proposal for the single
turnover event generating the cysteinyl-tyrosine thioether
cross-link. Each mechanism is discussed in the text (see
"Discussion").
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
18723-18733)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.H.Stipanuk,
C.R.Simmons,
P.Andrew Karplus,
and
J.E.Dominy
(2011).
Thiol dioxygenases: unique families of cupin proteins.
|
| |
Amino Acids,
41,
91.
|
 |
|
|
|
|
 |
M.H.Stipanuk,
and
I.Ueki
(2011).
Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur.
|
| |
J Inherit Metab Dis,
34,
17-32.
|
 |
|
|
|
|
 |
V.L.Davidson
(2011).
Generation of protein-derived redox cofactors by posttranslational modification.
|
| |
Mol Biosyst,
7,
29-37.
|
 |
|
|
|
|
 |
M.H.Stipanuk,
I.Ueki,
J.E.Dominy,
C.R.Simmons,
and
L.L.Hirschberger
(2009).
Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels.
|
| |
Amino Acids,
37,
55-63.
|
 |
|
|
|
|
 |
S.Leitgeb,
G.D.Straganz,
and
B.Nidetzky
(2009).
Functional characterization of an orphan cupin protein from Burkholderia xenovorans reveals a mononuclear nonheme Fe2+-dependent oxygenase that cleaves beta-diketones.
|
| |
FEBS J,
276,
5983-5997.
|
 |
|
|
|
|
 |
T.Kleffmann,
S.A.Jongkees,
G.Fairweather,
S.M.Wilbanks,
and
G.N.Jameson
(2009).
Mass-spectrometric characterization of two posttranslational modifications of cysteine dioxygenase.
|
| |
J Biol Inorg Chem,
14,
913-921.
|
 |
|
|
|
|
 |
A.L.Cechin,
M.Sinigaglia,
N.Lemke,
S.Echeverrigaray,
O.G.Cabrera,
G.A.Pereira,
and
J.C.Mombach
(2008).
Cupin: a candidate molecular structure for the Nep1-like protein family.
|
| |
BMC Plant Biol,
8,
50.
|
 |
|
|
|
|
 |
C.R.Simmons,
K.Krishnamoorthy,
S.L.Granett,
D.J.Schuller,
J.E.Dominy,
T.P.Begley,
M.H.Stipanuk,
and
P.A.Karplus
(2008).
A putative Fe2+-bound persulfenate intermediate in cysteine dioxygenase.
|
| |
Biochemistry,
47,
11390-11392.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Dominy,
J.Hwang,
S.Guo,
L.L.Hirschberger,
S.Zhang,
and
M.H.Stipanuk
(2008).
Synthesis of amino acid cofactor in cysteine dioxygenase is regulated by substrate and represents a novel post-translational regulation of activity.
|
| |
J Biol Chem,
283,
12188-12201.
|
 |
|
|
|
|
 |
M.H.Stipanuk,
J.E.Dominy,
I.Ueki,
and
L.L.Hirschberger
(2008).
Measurement of Cysteine Dioxygenase Activity and Protein Abundance.
|
| |
Curr Protoc Toxicol,
38,
6.15.1.
|
 |
|
|
|
|
 |
M.S.Rogers,
R.Hurtado-Guerrero,
S.J.Firbank,
M.A.Halcrow,
D.M.Dooley,
S.E.Phillips,
P.F.Knowles,
and
M.J.McPherson
(2008).
Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen.
|
| |
Biochemistry,
47,
10428-10439.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.K.Lee,
M.M.Whittaker,
and
J.W.Whittaker
(2008).
The electronic structure of the Cys-Tyr(*) free radical in galactose oxidase determined by EPR spectroscopy.
|
| |
Biochemistry,
47,
6637-6649.
|
 |
|
|
|
|
 |
C.A.Joseph,
and
M.J.Maroney
(2007).
Cysteine dioxygenase: structure and mechanism.
|
| |
Chem Commun (Camb),
(),
3338-3349.
|
 |
|
|
|
|
 |
G.N.Phillips,
B.G.Fox,
J.L.Markley,
B.F.Volkman,
E.Bae,
E.Bitto,
C.A.Bingman,
R.O.Frederick,
J.G.McCoy,
B.L.Lytle,
B.S.Pierce,
J.Song,
and
S.N.Twigger
(2007).
Structures of proteins of biomedical interest from the Center for Eukaryotic Structural Genomics.
|
| |
J Struct Funct Genomics,
8,
73-84.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

