 |
PDBsum entry 2acu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2acu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
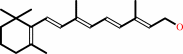 all-trans-retinol
all-trans-retinol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
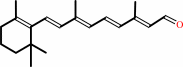 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
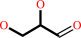 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
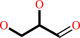 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
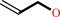 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
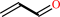 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:2021-2032
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme.
|
|
K.M.Bohren,
C.E.Grimshaw,
C.J.Lai,
D.H.Harrison,
D.Ringe,
G.A.Petsko,
K.H.Gabbay.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The active site of human aldose reductase contains two residues, His110 and
Tyr48, either of which could be the proton donor during catalysis. Tyr48 is a
candidate since its hydroxyl group is in proximity to Lys77 and thus may have an
abnormally low pKa value. To distinguish between these possibilities, we used
site-directed mutagenesis to create the H110Q and H110A, the Y48F, Y48H, and
Y48S, and the K77M mutant enzymes. The two His110 mutants resulted in a
1000-20,000-fold drop in kcat/Km, respectively, for the reduction of
DL-glyceraldehyde at pH 7. The Y48F mutation caused total loss of activity,
whereas the Y48H and Y48S mutants retained catalytic activity with kcat/Km
reduced by 5 orders of magnitude. The K77M mutant is an inactive enzyme. Kinetic
studies using xylose stereoisomers show that the wild-type enzyme distinguishes
between D-xylose, L-xylose, and D-lyxose up to 150-fold better than the H110A or
H110Q mutants. The His110 mutants do not effectively discriminate between these
isomers (4-11-fold). The crystal structure of the Y48H mutant refined at 1.8-A
resolution shows that the overall structure is not significantly different from
the wild-type structure. Electron densities for the histidine side chain and a
new water molecule fill the space occupied by Tyr48 in the wild-type enzyme. The
water molecule is in hydrogen-bonding distance to the N zeta group of Lys77 and
to the N epsilon of His48 and fills the space occupied by the hydroxyl group of
tyrosine in the wild-type structure. These findings suggest that proton transfer
is mediated in the Y48H mutant enzyme by the water molecule. The Y48H mutant
shows large and equal primary deuterium isotope effects on kcat and kcat/Km
(1.81 +/- 0.03), providing direct evidence for hydride transfer as the
rate-determining step in this mutant. Deuterium solvent isotope effects indicate
that the relative contribution of proton transfer to this step of the catalytic
cascade is much less important for the Y48H mutant than for the wild-type enzyme
[D2O(kcat/Km) = 1.06 +/- 0.02 and 4.73 +/- 0.23, respectively]. The kinetic and
mutagenesis data, together with structural data, indicate that His 110 plays an
important role in the orientation of substrates in the active site pocket, while
Tyr48 is the proton donor during aldehyde reduction by aldose reductase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.M.Katsori,
M.Chatzopoulou,
K.Dimas,
C.Kontogiorgis,
A.Patsilinakos,
T.Trangas,
and
D.Hadjipavlou-Litina
(2011).
Curcumin analogues as possible anti-proliferative & anti-inflammatory agents.
|
| |
Eur J Med Chem,
46,
2722-2735.
|
 |
|
|
|
|
 |
N.Nagata,
Y.Kusakari,
Y.Fukunishi,
T.Inoue,
and
Y.Urade
(2011).
Catalytic mechanism of the primary human prostaglandin F2α synthase, aldo-keto reductase 1B1--prostaglandin D2 synthase activity in the absence of NADP(H).
|
| |
FEBS J,
278,
1288-1298.
|
 |
|
|
|
|
 |
X.Chen,
Y.Yang,
B.Ma,
S.Zhang,
M.He,
D.Gui,
S.Hussain,
C.Jing,
C.Zhu,
Q.Yu,
and
Y.Liu
(2011).
Design and synthesis of potent and selective aldose reductase inhibitors based on pyridylthiadiazine scaffold.
|
| |
Eur J Med Chem,
46,
1536-1544.
|
 |
|
|
|
|
 |
D.H.Lee,
Y.J.Lee,
Y.W.Ryu,
and
J.H.Seo
(2010).
Molecular cloning and biochemical characterization of a novel erythrose reductase from Candida magnoliae JH110.
|
| |
Microb Cell Fact,
9,
43.
|
 |
|
|
|
|
 |
G.A.Khoury,
H.Fazelinia,
J.W.Chin,
R.J.Pantazes,
P.C.Cirino,
and
C.D.Maranas
(2009).
Computational design of Candida boidinii xylose reductase for altered cofactor specificity.
|
| |
Protein Sci,
18,
2125-2138.
|
 |
|
|
|
|
 |
B.Guillot,
C.Jelsch,
A.Podjarny,
and
C.Lecomte
(2008).
Charge-density analysis of a protein structure at subatomic resolution: the human aldose reductase case.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
567-588.
|
 |
|
|
|
|
 |
M.P.Blakeley,
F.Ruiz,
R.Cachau,
I.Hazemann,
F.Meilleur,
A.Mitschler,
S.Ginell,
P.Afonine,
O.N.Ventura,
A.Cousido-Siah,
M.Haertlein,
A.Joachimiak,
D.Myles,
and
A.Podjarny
(2008).
Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
|
| |
Proc Natl Acad Sci U S A,
105,
1844-1848.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.A.Barski,
S.M.Tipparaju,
and
A.Bhatnagar
(2008).
The aldo-keto reductase superfamily and its role in drug metabolism and detoxification.
|
| |
Drug Metab Rev,
40,
553-624.
|
 |
|
|
|
|
 |
S.M.Tipparaju,
O.A.Barski,
S.Srivastava,
and
A.Bhatnagar
(2008).
Catalytic mechanism and substrate specificity of the beta-subunit of the voltage-gated potassium channel.
|
| |
Biochemistry,
47,
8840-8854.
|
 |
|
|
|
|
 |
M.Biadene,
I.Hazemann,
A.Cousido,
S.Ginell,
A.Joachimiak,
G.M.Sheldrick,
A.Podjarny,
and
T.R.Schneider
(2007).
The atomic resolution structure of human aldose reductase reveals that rearrangement of a bound ligand allows the opening of the safety-belt loop.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
665-672.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Chang,
T.A.Griest,
T.M.Harter,
and
J.M.Petrash
(2007).
Functional studies of aldo-keto reductases in Saccharomyces cerevisiae.
|
| |
Biochim Biophys Acta,
1773,
321-329.
|
 |
|
|
|
|
 |
T.M.Penning,
and
J.E.Drury
(2007).
Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms.
|
| |
Arch Biochem Biophys,
464,
241-250.
|
 |
|
|
|
|
 |
J.M.Brownlee,
E.Carlson,
A.C.Milne,
E.Pape,
and
D.H.Harrison
(2006).
Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
|
| |
Bioorg Chem,
34,
424-444.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.I.Howard,
R.Sanishvili,
R.E.Cachau,
A.Mitschler,
B.Chevrier,
P.Barth,
V.Lamour,
M.Van Zandt,
E.Sibley,
C.Bon,
D.Moras,
T.R.Schneider,
A.Joachimiak,
and
A.Podjarny
(2004).
Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A.
|
| |
Proteins,
55,
792-804.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Obmolova,
A.Teplyakov,
P.P.Khil,
A.J.Howard,
R.D.Camerini-Otero,
and
G.L.Gilliland
(2003).
Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase.
|
| |
Proteins,
53,
323-325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.K.Lee,
B.S.Koo,
and
S.Y.Kim
(2003).
Cloning and characterization of the xyl1 gene, encoding an NADH-preferring xylose reductase from Candida parapsilosis, and its functional expression in Candida tropicalis.
|
| |
Appl Environ Microbiol,
69,
6179-6188.
|
 |
|
|
|
|
 |
S.D.Goldberg,
W.Iannuccilli,
T.Nguyen,
J.Ju,
and
V.W.Cornish
(2003).
Identification of residues critical for catalysis in a class C beta-lactamase by combinatorial scanning mutagenesis.
|
| |
Protein Sci,
12,
1633-1645.
|
 |
|
|
|
|
 |
U.Mura,
M.Cappiello,
P.G.Vilardo,
I.Cecconi,
M.Dal Monte,
and
A.Del Corso
(2003).
Signalling potential and protein modifying ability of physiological thiols.
|
| |
Biofactors,
17,
279-285.
|
 |
|
|
|
|
 |
C.R.Campomanes,
K.I.Carroll,
L.N.Manganas,
M.E.Hershberger,
B.Gong,
D.E.Antonucci,
K.J.Rhodes,
and
J.S.Trimmer
(2002).
Kv beta subunit oxidoreductase activity and Kv1 potassium channel trafficking.
|
| |
J Biol Chem,
277,
8298-8305.
|
 |
|
|
|
|
 |
E.Y.Jeong,
I.S.Kim,
and
H.Lee
(2002).
Identification of lysine-78 as an essential residue in the Saccharomyces cerevisiae xylose reductase.
|
| |
FEMS Microbiol Lett,
209,
223-228.
|
 |
|
|
|
|
 |
I.Cecconi,
A.Scaloni,
G.Rastelli,
M.Moroni,
P.G.Vilardo,
L.Costantino,
M.Cappiello,
D.Garland,
D.Carper,
J.M.Petrash,
A.Del Corso,
and
U.Mura
(2002).
Oxidative modification of aldose reductase induced by copper ion. Definition of the metal-protein interaction mechanism.
|
| |
J Biol Chem,
277,
42017-42027.
|
 |
|
|
|
|
 |
B.Nidetzky,
P.Mayr,
W.Neuhauser,
and
M.Puchberger
(2001).
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida tenuis.
|
| |
Chem Biol Interact,
130,
583-595.
|
 |
|
|
|
|
 |
E.Y.Jeong,
C.Sopher,
I.S.Kim,
and
H.Lee
(2001).
Mutational study of the role of tyrosine-49 in the Saccharomyces cerevisiae xylose reductase.
|
| |
Yeast,
18,
1081-1089.
|
 |
|
|
|
|
 |
J.M.Petrash,
B.S.Murthy,
M.Young,
K.Morris,
L.Rikimaru,
T.A.Griest,
and
T.Harter
(2001).
Functional genomic studies of aldo-keto reductases.
|
| |
Chem Biol Interact,
130,
673-683.
|
 |
|
|
|
|
 |
M.J.Ondrechen,
J.G.Clifton,
and
D.Ringe
(2001).
THEMATICS: a simple computational predictor of enzyme function from structure.
|
| |
Proc Natl Acad Sci U S A,
98,
12473-12478.
|
 |
|
|
|
|
 |
V.Nahoum,
A.Gangloff,
P.Legrand,
D.W.Zhu,
L.Cantin,
B.S.Zhorov,
V.Luu-The,
F.Labrie,
R.Breton,
and
S.X.Lin
(2001).
Structure of the human 3alpha-hydroxysteroid dehydrogenase type 3 in complex with testosterone and NADP at 1.25-A resolution.
|
| |
J Biol Chem,
276,
42091-42098.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Lamotte-Brasseur,
A.Dubus,
and
R.C.Wade
(2000).
pK(a) calculations for class C beta-lactamases: the role of Tyr-150.
|
| |
Proteins,
40,
23-28.
|
 |
|
|
|
|
 |
J.M.Jez,
J.L.Ferrer,
M.E.Bowman,
R.A.Dixon,
and
J.P.Noel
(2000).
Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
|
| |
Biochemistry,
39,
890-902.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Ye,
D.Hyndman,
X.Li,
T.G.Flynn,
and
Z.Jia
(2000).
Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
|
| |
Proteins,
38,
41-48.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Rink,
J.Kingma,
J.H.Lutje Spelberg,
and
D.B.Janssen
(2000).
Tyrosine residues serve as proton donor in the catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacter.
|
| |
Biochemistry,
39,
5600-5613.
|
 |
|
|
|
|
 |
S.Khurana,
G.Sanli,
D.B.Powers,
S.Anderson,
and
M.Blaber
(2000).
Molecular modeling of substrate binding in wild-type and mutant Corynebacteria 2,5-diketo-D-gluconate reductases.
|
| |
Proteins,
39,
68-75.
|
 |
|
|
|
|
 |
V.Calderone,
B.Chevrier,
M.Van Zandt,
V.Lamour,
E.Howard,
A.Poterszman,
P.Barth,
A.Mitschler,
J.Lu,
D.M.Dvornik,
G.Klebe,
O.Kraemer,
A.R.Moorman,
D.Moras,
and
A.Podjarny
(2000).
The structure of human aldose reductase bound to the inhibitor IDD384.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
536-540.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Lefrançois-Martinez,
C.Tournaire,
A.Martinez,
M.Berger,
S.Daoudal,
D.Tritsch,
G.Veyssière,
and
C.Jean
(1999).
Product of side-chain cleavage of cholesterol, isocaproaldehyde, is an endogenous specific substrate of mouse vas deferens protein, an aldose reductase-like protein in adrenocortical cells.
|
| |
J Biol Chem,
274,
32875-32880.
|
 |
|
|
|
|
 |
J.M.Gulbis,
S.Mann,
and
R.MacKinnon
(1999).
Structure of a voltage-dependent K+ channel beta subunit.
|
| |
Cell,
97,
943-952.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.M.Hevel,
S.A.Mills,
and
J.P.Klinman
(1999).
Mutation of a strictly conserved, active-site residue alters substrate specificity and cofactor biogenesis in a copper amine oxidase.
|
| |
Biochemistry,
38,
3683-3693.
|
 |
|
|
|
|
 |
L.Costantino,
G.Rastelli,
P.Vianello,
G.Cignarella,
and
D.Barlocco
(1999).
Diabetes complications and their potential prevention: aldose reductase inhibition and other approaches.
|
| |
Med Res Rev,
19,
3.
|
 |
|
|
|
|
 |
P.J.Oates,
and
B.L.Mylari
(1999).
Aldose reductase inhibitors: therapeutic implications for diabetic complications.
|
| |
Expert Opin Investig Drugs,
8,
2095-2119.
|
 |
|
|
|
|
 |
P.Várnai,
W.G.Richards,
and
P.D.Lyne
(1999).
Modelling the catalytic reaction in human aldose reductase.
|
| |
Proteins,
37,
218-227.
|
 |
|
|
|
|
 |
T.Suzuki,
Y.Fujii,
M.Miyano,
L.Y.Chen,
T.Takahashi,
and
K.Watanabe
(1999).
cDNA cloning, expression, and mutagenesis study of liver-type prostaglandin F synthase.
|
| |
J Biol Chem,
274,
241-248.
|
 |
|
|
|
|
 |
B.P.Schlegel,
J.M.Jez,
and
T.M.Penning
(1998).
Mutagenesis of 3 alpha-hydroxysteroid dehydrogenase reveals a "push-pull" mechanism for proton transfer in aldo-keto reductases.
|
| |
Biochemistry,
37,
3538-3548.
|
 |
|
|
|
|
 |
B.P.Schlegel,
K.Ratnam,
and
T.M.Penning
(1998).
Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine.
|
| |
Biochemistry,
37,
11003-11011.
|
 |
|
|
|
|
 |
D.Cao,
S.T.Fan,
and
S.S.Chung
(1998).
Identification and characterization of a novel human aldose reductase-like gene.
|
| |
J Biol Chem,
273,
11429-11435.
|
 |
|
|
|
|
 |
H.Lee
(1998).
The structure and function of yeast xylose (aldose) reductases.
|
| |
Yeast,
14,
977-984.
|
 |
|
|
|
|
 |
J.M.Jez,
and
T.M.Penning
(1998).
Engineering steroid 5 beta-reductase activity into rat liver 3 alpha-hydroxysteroid dehydrogenase.
|
| |
Biochemistry,
37,
9695-9703.
|
 |
|
|
|
|
 |
M.J.Crabbe,
and
D.Goode
(1998).
Aldose reductase: a window to the treatment of diabetic complications?
|
| |
Prog Retin Eye Res,
17,
313-383.
|
 |
|
|
|
|
 |
S.T.Kim,
W.K.Huh,
B.H.Lee,
and
S.O.Kang
(1998).
D-arabinose dehydrogenase and its gene from Saccharomyces cerevisiae.
|
| |
Biochim Biophys Acta,
1429,
29-39.
|
 |
|
|
|
|
 |
W.Neuhauser,
D.Haltrich,
K.D.Kulbe,
and
B.Nidetzky
(1998).
Noncovalent enzyme-substrate interactions in the catalytic mechanism of yeast aldose reductase.
|
| |
Biochemistry,
37,
1116-1123.
|
 |
|
|
|
|
 |
Y.S.Lee,
M.Hodoscek,
B.R.Brooks,
and
P.F.Kador
(1998).
Catalytic mechanism of aldose reductase studied by the combined potentials of quantum mechanics and molecular mechanics.
|
| |
Biophys Chem,
70,
203-216.
|
 |
|
|
|
|
 |
M.J.Bennett,
R.H.Albert,
J.M.Jez,
H.Ma,
T.M.Penning,
and
M.Lewis
(1997).
Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
|
| |
Structure,
5,
799-812.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.el-Kabbani,
D.A.Carper,
M.H.McGowan,
Y.Devedjiev,
K.J.Rees-Milton,
and
T.G.Flynn
(1997).
Studies on the inhibitor-binding site of porcine aldehyde reductase: crystal structure of the holoenzyme-inhibitor ternary complex.
|
| |
Proteins,
29,
186-192.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.H.Walker,
and
N.C.Bruce
(1996).
Towards engineering an improved morphine dehydrogenase.
|
| |
Ann N Y Acad Sci,
799,
6.
|
 |
|
|
|
|
 |
K.Kita,
K.Matsuzaki,
T.Hashimoto,
H.Yanase,
N.Kato,
M.C.Chung,
M.Kataoka,
and
S.Shimizu
(1996).
Cloning of the aldehyde reductase gene from a red yeast, Sporobolomyces salmonicolor, and characterization of the gene and its product.
|
| |
Appl Environ Microbiol,
62,
2303-2310.
|
 |
|
|
|
|
 |
M.J.Bennett,
B.P.Schlegel,
J.M.Jez,
T.M.Penning,
and
M.Lewis
(1996).
Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
|
| |
Biochemistry,
35,
10702-10711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.von Itzstein,
and
P.Colman
(1996).
Design and synthesis of carbohydrate-based inhibitors of protein-carbohydrate interactions.
|
| |
Curr Opin Struct Biol,
6,
703-709.
|
 |
|
|
|
|
 |
O.A.Barski,
K.H.Gabbay,
and
K.M.Bohren
(1996).
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity.
|
| |
Biochemistry,
35,
14276-14280.
|
 |
|
|
|
|
 |
P.M.Kiefer,
K.I.Varughese,
Y.Su,
N.H.Xuong,
C.F.Chang,
P.Gupta,
T.Bray,
and
J.M.Whiteley
(1996).
Altered structural and mechanistic properties of mutant dihydropteridine reductases.
|
| |
J Biol Chem,
271,
3437-3444.
|
 |
|
|
|
|
 |
R.L.Kingston,
R.K.Scopes,
and
E.N.Baker
(1996).
The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP.
|
| |
Structure,
4,
1413-1428.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Nakano,
and
J.M.Petrash
(1996).
Kinetic and spectroscopic evidence for active site inhibition of human aldose reductase.
|
| |
Biochemistry,
35,
11196-11202.
|
 |
|
|
|
|
 |
D.A.Carper,
T.C.Hohman,
and
S.E.Old
(1995).
Residues affecting the catalysis and inhibition of rat lens aldose reductase.
|
| |
Biochim Biophys Acta,
1246,
67-73.
|
 |
|
|
|
|
 |
D.H.Harrison
(1995).
All in the family.
|
| |
Nat Struct Biol,
2,
719-720.
|
 |
|
|
|
|
 |
H.M.Wilks,
and
M.P.Timko
(1995).
A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: identification and mutagenesis of two conserved residues that are essential for enzyme activity.
|
| |
Proc Natl Acad Sci U S A,
92,
724-728.
|
 |
|
|
|
|
 |
O.el-Kabbani,
K.Judge,
S.L.Ginell,
D.A.Myles,
L.J.DeLucas,
and
T.G.Flynn
(1995).
Structure of porcine aldehyde reductase holoenzyme.
|
| |
Nat Struct Biol,
2,
687-692.
|
 |
|
|
|
|
 |
T.J.Kubiseski,
and
T.G.Flynn
(1995).
Studies on human aldose reductase. Probing the role of arginine 268 by site-directed mutagenesis.
|
| |
J Biol Chem,
270,
16911-16917.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

